1. Background
Staphylococcus aureus is one of the most important pathogens for humans and the second most common cause of nosocomial infections (1, 2). Staphylococcal infections occur continuously in hospitalized patients and have severe consequences ranging from superficial skin and soft tissue infections to severe systematic infections (3). The increasing trend of antibiotic-resistant strains of S. aureus, mostly methicillin-resistant S. aureus (MRSA), is a significant concern toward the management of related diseases (4, 5). Methicillin-resistant S. aureus is an important cause of hospital-acquired infections because of rapidly adapting to selective pressures encountered in hospital environments. Understanding antibiotic resistance mechanisms can be useful to overcome the emergence of methicillin-sensitive S. aureus (MSSA)strains (6).
The transfer of antibiotic resistance genes by horizontal gene transfer promotes antibiotic resistance among bacteria, which, in turn, complicates the treatment processes of the infections (7). Antibiotic resistance genes are located on the various types of DNA mobilizing elements, such as plasmids, transposons, integrons, or phages (8, 9). Integrons are one of the key systems and primary factors involved in the transfer of multidrug resistance elements among bacteria. Integrons selectively receive and remove various drug resistance gene packages and transfer them to other bacterial strains, which, in fact, accelerates the transmission of drug resistance distribution (10, 11).
The important compositions of an integron element include the intI gene encoding a site-specific recombinase according to the integrase family and an adjacent site named att, which is identified by integrase and is a receptor for gene cassettes (10). The dispersion of antibiotic resistance genes due to the presence of integrons has become a major problem in treating infections caused by pathogenic bacteria (11). In this regard, their importance has also been noted in the dissemination of antibiotic resistance among Gram-positive pathogens, particularly S. aureus (12).
2. Objectives
Because of the importance of the presence of integrons as the main option for facilitating the transfer of antibiotic resistance genes in bacteria and shortcomings in reported papers from our region, the present study aimed to evaluate the presence of integrons and their relationships with antibiotic resistance in clinical isolates of S. aureus isolated from hospitalized patients.
3. Methods
3.1. Study Design and Bacterial Isolation
In this cross-sectional study during the first six months of 2017, a total of 134 clinical isolates of S. aureus from two major hospitals of Nemazee and Ghotboddin Shirazi were collected from different clinical specimens. Colonies were characterized as S. aureus by standard microbiological tests including positive reactions of catalase, Dnase, tube coagulase, and mannitol fermentation on the Mannitol salt agar. Then, all the isolates were confirmed by the mecA gene-based polymerase chain reaction (PCR) (13). The identified isolates were kept at -80ºC for further experiments.
3.2. Antibiotic Susceptibility Test
Staphylococcus aureus isolates were evaluated for antibiotic susceptibility by the disk diffusion technique on Mueller-Hinton agar according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (14). The results were documented after incubation for 16 - 18 hours at 35ºC. The used antibiotics included tetracycline: T (30 μg), rifampin: RP (5 μg), tobramycin: TN (10 μg), cefoxitin: CX (30 μg), gentamicin: Gm (10 μg), erythromycin: E (15 μg), clindamycin: CD (2 μg), ciprofloxacin: CIP (5 μg), and sulfamethoxazole-trimethoprim: SXT (30 μg). The breakpoints for antibiotic susceptibility were determined based on the CLSI recommendation.
3.3. Genomic DNA Extraction
The phenol-chloroform method previously introduced by Sambrook and Russell was used for whole genomic DNA extraction (15). The extracted DNA was washed with 70% ethanol and dissolved in 50 μL of TE buffer (10 mM Tris; 1 mM EDTA, pH 8). The solution was ultimately stored at -20ºC for storage. Finally, 1 μL of stored DNA was used as a template in the PCR.
3.4. Identification of Staphylococcus aureus
The phenotypic characterized S. aureus and MRSA were confirmed molecularly by the detection of the mecA gene using primers described in the study by Zhang et al. (13). The amplification of DNA was performed using thermal cycler 5530 (Eppendorf master, Germany) in a total volume of 25 μL in the following condition: 5 min at 95ºC for initial denaturation, followed by 35 cycles of denaturation at 94ºC for 60 s, annealing for 45 s (varying based on the sequences of primers), 1 min at 72ºC for extension, and 5 min at 72ºC for final extension. Electrophoresis of amplicons was used by the 1.5% agarose gel prepared in 1X TAE (Tris/Acetate/EDTA) buffer and visualized through UV transillumination after staining with safe stain load dye (CinnaGen Co., Iran).
3.5. Identification of Integrons
All isolates were tested to identify class 1-3 integrons using primers described in the study by Barraud et al. designed for the intI genes (16). The amplification of DNA was performed using the method described in section 3.4.
3.6. Statistical Analysis
The chi-square or Fisher exact test was performed to analyze the results as appropriate by SPSS software, version 21.0 (IBM Corp., USA). The categorical results were expressed as the percentages of the group in terms of relative frequency. A P value of < 0.05 was considered statistically significant.
4. Results
Totally, during the study period, 134 isolates of S. aureus from different wards of the study hospitals were collected and all of them were identified as S. aureus by phenotypic detection and the presence of the eap gene. Out of 134 S. aureus isolates included in our study, 80 (59.7%) and 54 (40.3%) isolates were obtained from male and female patients, respectively. The most frequent source of bacterial isolation was from respiratory tract infections (RTIs; 37.3%), followed by bloodstream infection (BSI; 23.9%), skin and soft tissue infections (SSTIs; 21.6%), urinary tract infection (UTI; 11.9%), eye infection (2.2%), abdominal infection (2.2%), and meningitis (0.7%). Based on the results of antibiotic susceptibility testing, 58 (43.2%) of 134 isolates of S. aureus were reported as MRSA using cefoxitin disc diffusion and mecA gene detection. The highest resistance was seen to tetracycline (48.5%) and the lowest to clindamycin (41.8%). The full results of the antibiotic susceptibility pattern of isolates classified by MRSA and MSSA are shown in Table 1.
| Antibiotic | MRSA | MSSA | P Value | Total |
|---|---|---|---|---|
| Gentamicin | 79.3 | 23.7 | < 0.001 | 47.8 |
| Tobramycin | 69 | 26.3 | 0.008 | 44.8 |
| Tetracycline | 55.2 | 43.4 | < 0.001 | 48.5 |
| Rifampin | 77.6 | 18.4 | < 0.001 | 44.0 |
| Erythromycin | 55.2 | 32.9 | 0.01 | 42.5 |
| Clindamycin | 44.8 | 39.5 | 0.029 | 41.8 |
| Ciprofloxacin | 70.7 | 22.4 | < 0.001 | 43.3 |
| Co-trimoxazole | 72.4 | 26.3 | 0.017 | 46.3 |
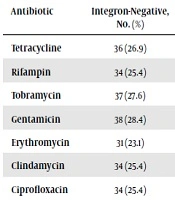
Molecular analysis showed that 53 (39.6%) and 5 (3.7%) S. aureus isolates were positive for class 1 and 2 integrons, respectively. Meanwhile, none of the isolates contained class 3 integron encoding genes. Meanwhile, 5 (3.7%) isolates (four from RTIs and one from UTI) contained both int1 and intI2 genes. The distribution of MRSA strains and S. aureus isolates containing class 1 and 2 integrons in different infection sources is presented in Table 2. The number and percentage of positive and negative strains in terms of the presence of class 1 integron in strains with antibiotic resistance, individually for each antibiotic, are shown in Table 3.
| Infection Source (N) | MRSA, % | Class 1 Integrons, % | Class 2 Integrons, % |
|---|---|---|---|
| Respiratory tract infection (50) | 46 | 48 | 8 |
| Bloodstream infection (32) | 50 | 18.8 | 0 |
| Skin and soft tissue infection (29) | 31 | 44.8 | 0 |
| Urinary tract infection (16) | 50 | 50 | 6.3 |
| Eye infection (3) | 33.3 | 33.3 | 0 |
| Abdominal infection (3) | 33.3 | 33.3 | 0 |
| Meningitis (1) | 0 | 0 | 0 |
| Antibiotic | Integron-Negative, No. (%) | Integron-Positive, No. (%) | P Value |
|---|---|---|---|
| Tetracycline | 36 (26.9) | 29 (21.7) | 0.789 |
| Rifampin | 34 (25.4) | 25 (18.7) | < 0.001 |
| Tobramycin | 37 (27.6) | 24 (17.9) | < 0.001 |
| Gentamicin | 38 (28.4) | 26 (19.4) | 0.059 |
| Erythromycin | 31 (23.1) | 26 (19.4) | 0.358 |
| Clindamycin | 34 (25.4) | 22 (16.4) | 0.003 |
| Ciprofloxacin | 34 (25.4) | 24 (17.9) | 0.001 |
| Co-trimoxazole | 40 (29.9) | 22 (16.4) | 0.001 |
5. Discussion
In this hospital-based survey, we investigated the occurrence of prevalent integrons in clinical isolates of S. aureus isolated from two major hospitals in southwestern Iran. Staphylococcus aureus, particularly MRSA, is one of the most important human pathogenic bacteria (17). With the introduction of various antibiotics, the mortality rate of infections caused by this bacterium was reduced, but nowadays, this bacterium is becoming resistant to many current antibiotics by improving its resistance mechanisms (17). The transfer rate of drug resistance genes by using horizontal gene transfer can speed up the process of displacement of drug resistance genes (18, 19). Our findings indicated that 43.2% of all samples obtained from different wards of hospitals were MRSA isolates. In this regard, 35.2% of S. aureus isolates examined in a study in Ahvaz (2016), southwestern Iran, were considered as MRSA (20). The MRSA rate in another study performed among burn patients in Ahvaz (2012) was estimated to be 86.4% that compared to our study showed a much higher value (21). In a study conducted in China in 2009, from 186 isolates of S. aureus, 67.9% were MRSA (22). These differences may be due to variations in geographical distribution, sample size, and source of infections.
In the present study, the resistance pattern of nine locally available antibiotics against clinical isolates of S. aureus was determined. The overall resistance rates of the tested antibiotics were almost the same; however, among MRSA isolates, the most effective agent was clindamycin, followed by erythromycin and tetracycline. Among MSSA isolates, the most effective agents were rifampicin, followed by ciprofloxacin. Therefore, these options can be recommended for the management of staphylococcal infections in our region by considering the cost-effectiveness of the used antibiotics. Although the antibiotic resistance pattern of S. aureus can be varied in different geographic regions or even within hospitals in the same region, there are several results in Iran and other countries comparable with our findings (22-24).
To best of our knowledge, this study for the first time reports the occurrence of class 1 integrons (39.6%) and class 2 integrons (3.7%) in clinical isolates of S. aureus in Shiraz, southwestern Iran. Integrons not commonly occur in Gram-positive bacteria, but integrons with the ability to transmit resistance determinants have a high capacity for dissemination within bacteria. Therefore, any effort to determine the burden of integrons-positive bacteria has a vital role in the blockage of the emergence and spread of antibiotic-resistant strains (12). The diverse nature of integrons among different types of Gram-positive bacteria or sources in the literature is almost undeniable, but as a general concept from those studies, class 1 integron is the predominant type of integrons that is likely to play a role in antibiotic resistance of resistant strains (2, 12, 23-25).
5.1. Conclusions
As far as we know, this study is the first study that showed the prevalence of integrons in S. aureus in Shiraz, southwestern Iran. On the other hand, the information obtained from this study shows the high number of positive strains for integrons in nosocomial isolates of S. aureus. These data alone cannot confirm the association between integrons and the presence of antibiotic resistance genes. Furthermore, our findings highlight the relevance of investigating the spread of drug resistance genes in S. aureus for controlling its dissemination through the hospital environments.