1. Background
Leishmaniasis are ancient neglected tropical diseases (NTDs) caused by over 21 species of intracellular Leishmania parasites with sand flies as vectors and a broad range of clinical presentations comprising simple, diffuse and mucocutaneous leishmaniasis, as well as the visceral disease (1). Approximately, 100 countries are located in leishmaniasis endemic regions and 350 million people are at risk of infection (2). Cutaneous leishmaniasis and visceral disease are two important manifestations of leishmaniasis with the incidence of 0.7 - 1.2 and 0.2 - 0.4 million new cases per year, respectively (3).
Cutaneous leishmaniasis is most highlighted in 10 nations including Columbia, Brazil, Costa Rica, Peru, Iran, Syria, Afghanistan, Algeria, Ethiopia, and North Sudan (2). A number of risk factors such as migration to endemic areas, urbanization with the increased risk of vector localization and man-made manipulation in the environment are involved in the occurrence and distribution of cutaneous leishmaniasis (4). Countries located in the Eastern Mediterranean territory such as Iran possess appropriate conditions for cutaneous leishmaniasis endemicity (5). Although neglected, it is estimated that 30.9 new cutaneous leishmaniasis cases per 100000 population occur in Iran, which is mostly highlighted in Isfahan and Fars provinces (3). After malaria, cutaneous leishmaniasis is the second most prevalent arthropod-borne disease in the country, representing as two major forms: (1) anthroponotic, dry, or urban leishmaniasis due to Leishmania tropica particularly in cities with high population density and daily immigrants and (2) zoonotic, wet, or rural leishmaniasis owing to L. major occurring in rustic areas with weak hygienic infrastructures and rodent species as reservoir hosts (3).
Routine parasitology examination for the diagnosis of cutaneous leishmaniasis is lesion sampling and observation of intracellular amastigote forms in macrophages. However, such a technique does not detect the exact species and/or strains of the parasite for better subsequent treatment options and prevention strategies (6). Hence, sophisticated molecular tests are recommended for species identification and detection of resistant isolates. The highly-conserved fragment of internal transcribed spacer-1 (ITS1) from ribosomal RNA (rRNA) is a classic genetic hallmark for phylogenetic categorization of closely-related species. The parasite species/strains and their likely mutant forms can be determined using restriction enzymes in polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) in the ITS1 region (7, 8).
So far, several studies have been conducted on the genotyping of cutaneous leishmaniasis agents in various geographical areas of Iran. For instance, Kermanjani et al. proved all clinical isolates as L. major using HaeIII enzyme digestion, without strain and mutation analysis (9). Another genotyping study of clinical isolates in Isfahan and Bam by HaeIII and TaqI enzyme digestion showed the presence of LmA and LmB as L. major strains, while LtA and LtB were the specified strains of L. tropica (10). A considerable part of our knowledge of the genomic structure of cutaneous leishmaniasis species in Iran originates from studies performed in western/southwestern endemic foci. Previous studies carried out retrospective investigations, comparable culture, and mini-exon PCR, as well as PCR-RFLP for species determination, in this endemic territory (11-13).
2. Objectives
The current original study was accomplished to assess the molecular identification of cutaneous leishmaniasis species/strains with emphasis on possible mutations in clinical samples from suspected patients in Dasht-e-Azadegan county, Khuzestan, Southwestern Iran.
3. Methods
3.1. Study Population and Samples
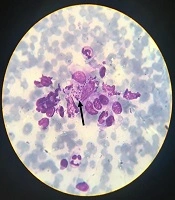
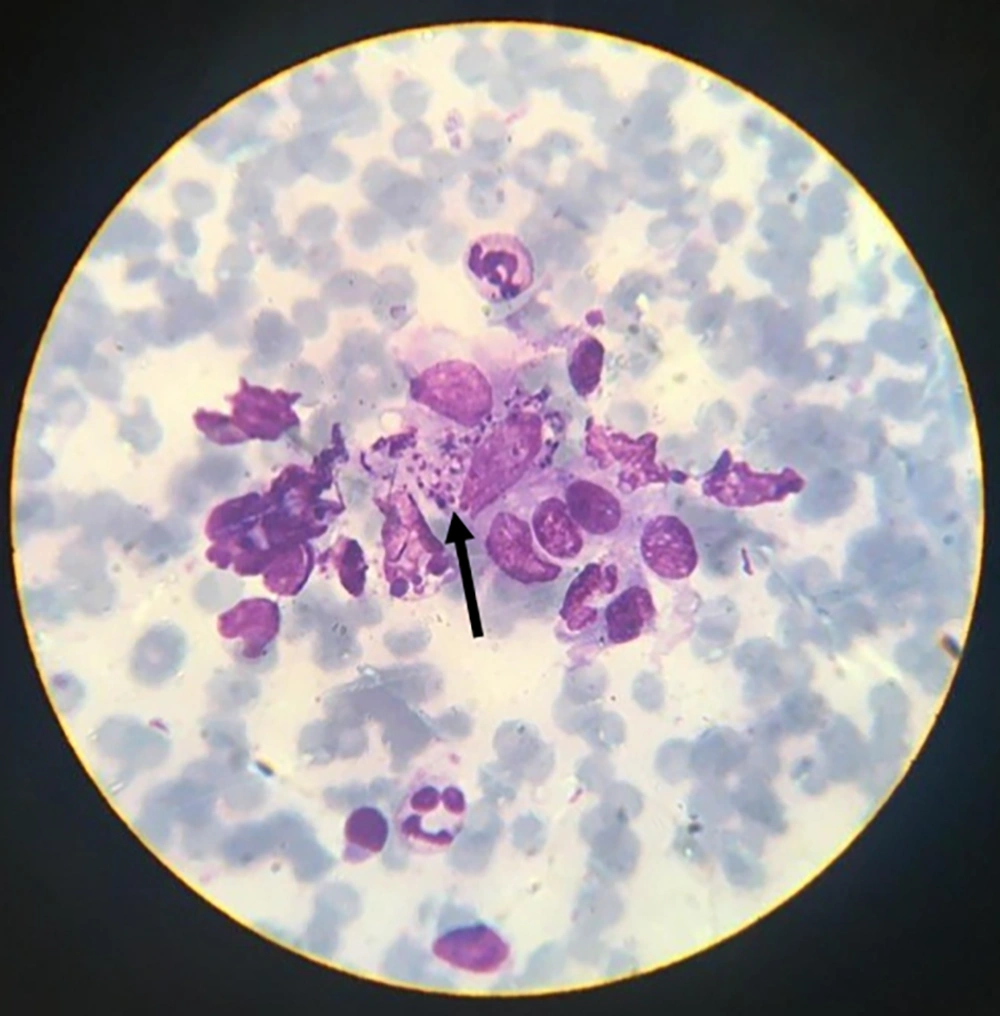
In this study, 80 samples were obtained as slide smears from dermal lesions of suspected individuals referring to Dasht-e-Azadegan health centers, Southwest of Iran, in 2016 and confirmed by the centers. All specimens were accurately examined by Giemsa staining and light microscopy to observe amastigote forms of genus Leishmania and confirm cutaneous leishmaniasis.
3.2. DNA Extraction and PCR-RFLP Technique
Following washing the slides with PBS solution, the smear contents were scraped using a scalpel and transferred to a 1.5 mL microtube. After a second PBS washing, the GeNet Bio PrimePrep DNA extraction kit from tissue (Genet Bio, South Korea) was used for DNA extraction, based on the manufacturer’s protocol. Finally, the extracted DNA material was kept at -20°C for next use.
ITS1-PCR was applied using the LITSR (5-CTGGATCATTTTCCGATG-3) and L5.8S (5-TGATACCACTTATCGCACTT-3) primer pair to detect genus Leishmania in clinical isolates. For this aim, the 25 µL reaction mixture consisted of 1.5 mM MgCl2, 250 µM dNTPs, 1 U Taq DNA polymerase, and 10 pmol of each primer. The amplification process was done in a thermal cycler (Analytik Jena, Germany) in the following condition: initial denaturation at 95°C for 5 minutes, 35 cycles of denaturation at 94°C for 30 seconds, annealing at 55°C for 45 seconds, extension at 72°C for 45 seconds; and final extension at 72°C for 5 minutes. Following genus determination by PCR, species/strains were identified using the RFLP method (14). Accordingly, Leishmania species were discerned by HaeIII (Jena Bioscience, Germany) with 5 µL PCR product, 2.5 µL universal buffer, 1.5 µL enzyme, and 6 µL sterile deionized water.
Strains were distinguished by TaqI (Jena Bioscience, Germany) under the previous reaction condition. Moreover, DpnI (Thermo Fisher Scientific, USA) and HpaII (Jena Bioscience, Germany) were used for mutation analysis based on a reaction containing 6 µL PCR product, 2 µL universal buffer, 1 µL enzyme, and 7 µL sterile molecular-grade water. All mixtures were incubated at 37°C for 3 hours, except for the TaqI mixture that was incubated at 65°C for 3 hours. The 1.5% and 2% gel electrophoresis, followed by gel-doc monitoring, was applied for PCR and digestion products, respectively.
4. Results
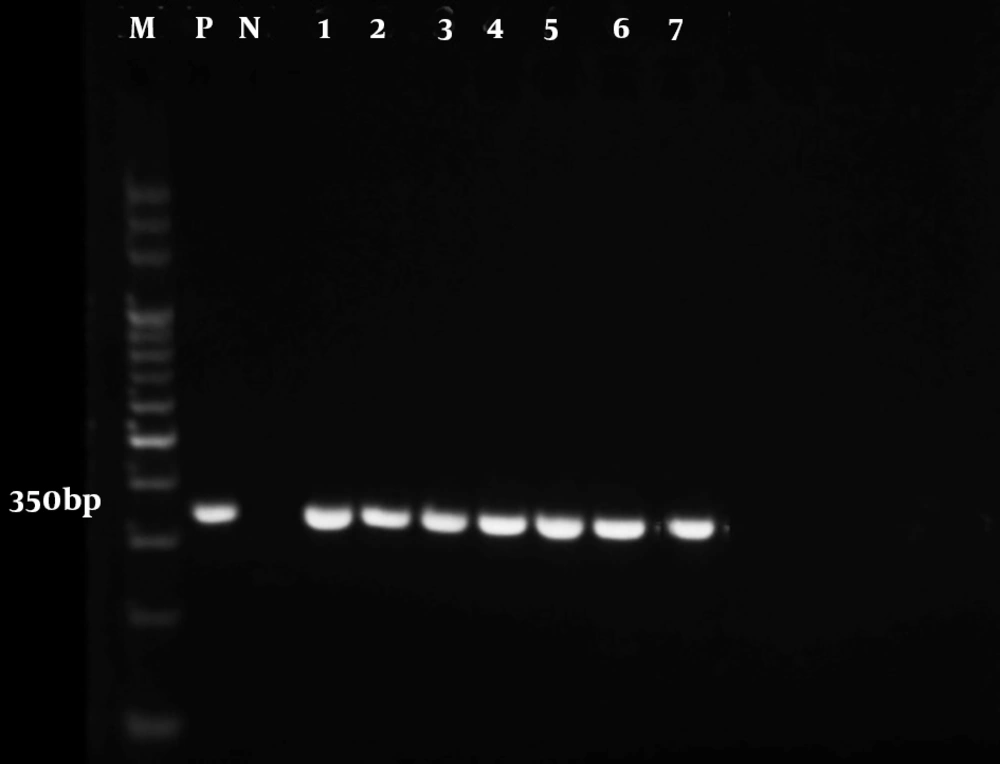
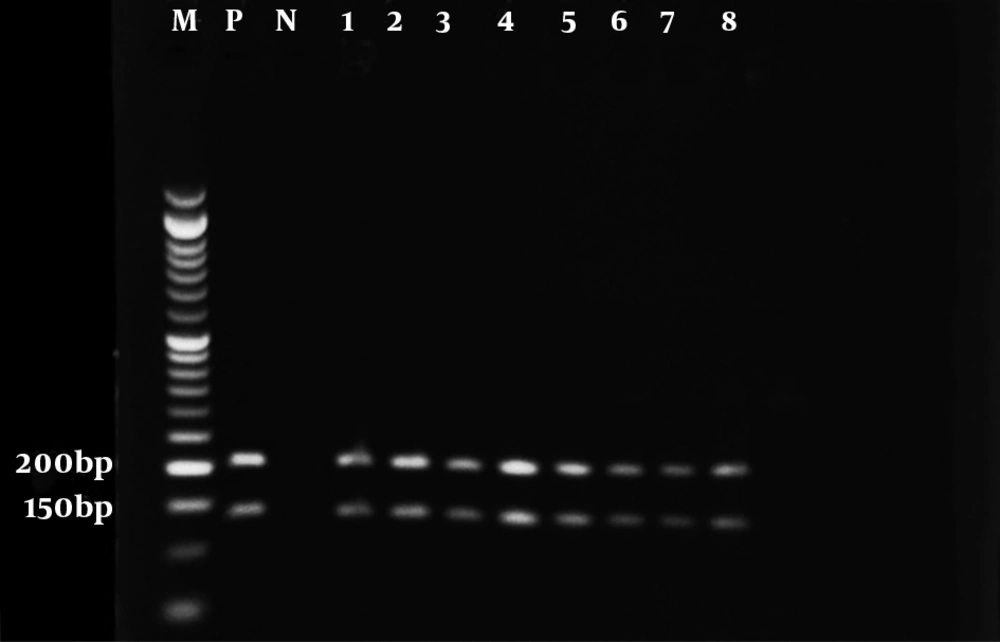
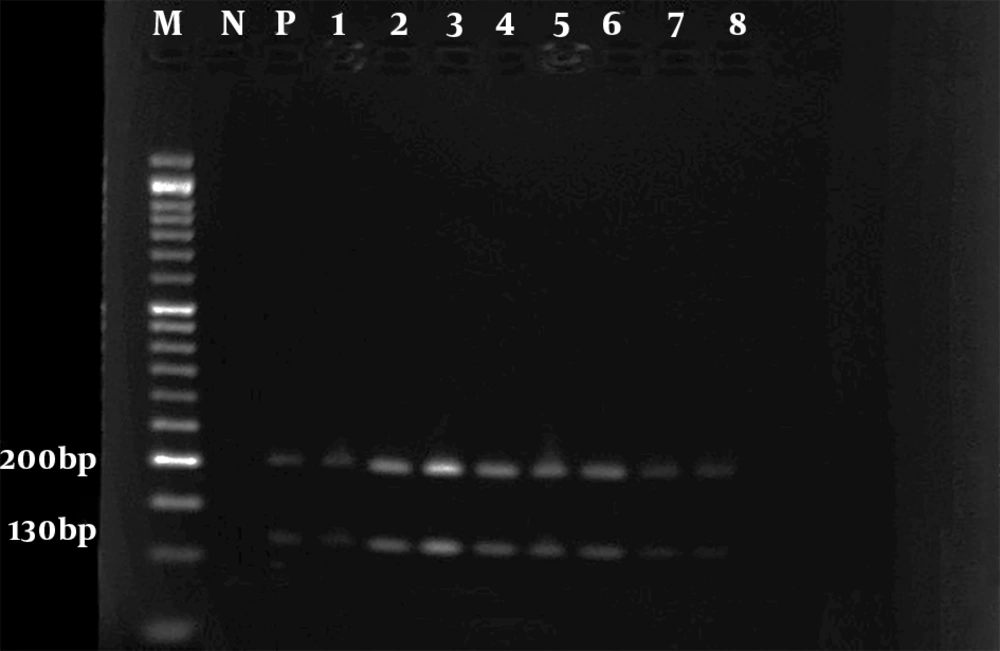
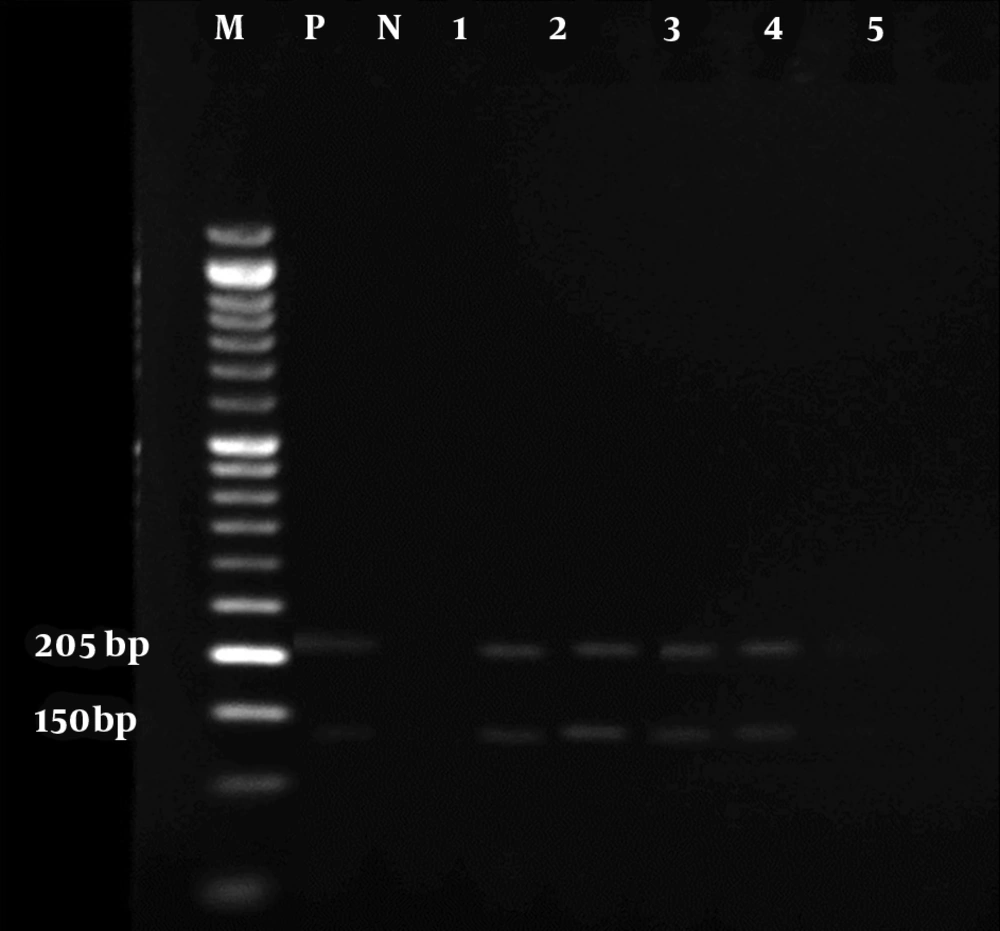
The results of the microscopic examination indicated that all the 80 suspected slide smears collected in this study were confirmed as cutaneous leishmaniasis (Figure 1). The PCR technique demonstrated genus Leishmania in all specimens by representing a 350 bp band in accordance with the amplification of the ITS1 region (Figure 2). On the basis of HaeIII digestion, all samples showed 150 and 200 bp patterns, representing L. major species (Figure 3). Then, TaqI digestion rendered A1 strain of L. major (LmA1) with 130 and 200 bp band patterns (Figure 4). Mutation analysis was carried out by DpnI rendering 150 and 205 bp bands on PCR products (Figure 5). Moreover, the application of HpaII restriction enzyme caused no digestion; thus, no mutation was found in the ITS1 fragment of the isolated species.
5. Discussion
Leishmaniases are NTDs being endemic in 98 countries with over 350 million at-risk individuals. However, the occurrence of this infection is more than expected, due to under-diagnosis and/or non-compulsory infection report (6). Previously, the diagnosis was mostly based on traditional identification of lesions, as well as biological classification and isoenzyme typing. More recently, unprecedented nucleic acid-based techniques have facilitated the determination of Leishmania species/strains (8). The cutaneous disease due to Leishmania is the most prevalent form with an incidence rate of 20000 people annually in Iran (3). The current study aimed to address the species/strains of cutaneous leishmaniasis causative agents in people referring to the health centers of Dasht-e-Azadegan, Khuzestan province, using ITS1 PCR-RFLP in 2016. According to our findings, all samples showed a 350 bp band following ITS1-PCR, suggesting the molecular identification of genus Leishmania by the LITSR and L5.8S primer pair, which is consistent to Schonian et al research (15). Similarly, most studies in Iran have used this primer pair for the molecular detection of Leishmania (9, 16-20).
However, Mohammadiha et al. also used another ITS-specific primer pair yielding 800 - 850 bp PCR product (21). Next, species-dependent digestion using HaeIII produced 150 and 200 bp bands belonging to L. major. TaqI digestion revealed the A1 strain of L. major with digestion range of 120 to 210 bp. Moreover, no mutation site was determined in the obtained isolates by DpnI and HpaII digestion (22). Thus, all 80 samples isolated from suspected cases were identified by PCR-RFLP as LmA1 strains. In a similar investigation in Ilam province, Kermanjani et al. determined all microscopically confirmed clinical isolates as L. major; however, strains and mutations were not analyzed (9).
Many studies have documented L. major and other species in Iran and around the globe. For instance, Mahmoudzadeh-Niknam et al. found 341 Leishmania isolates including 58 L. tropica isolates and 283 L. major isolates in 11 geographical territories of Iran (23). Another study in Ahvaz, Dasht-e-Azadegan, Shush, Hendijan, and Ramhormoz revealed 14 Leishmania samples with 2 L. tropica isolates and 12 L. major isolates (13). According to Maraghi et al. 90% of the isolates from Shush county were L. major, while the remaining 10% were L. tropica (24). Based on previous studies in the province, it could be expected that L. major is the predominant species in the area. On the other hand, the traits of Tatera indica (reservoir) and Phlebotomus papatasi (vector) in the area, as well as various documentations from Khuzestan province, implicate this province along with Ilam and Bushehr provinces as the most highlighted regions for zoonotic cutaneous leishmaniasis in Iran (3, 25-27).
Mohammadiha et al. amplified ITS1 by MO-F and MO-R primers and made digestion using TaqI, yielding L. major (33 cases) and L. tropica (61 cases) in all clinical samples. They could also identify L. tropica (43 cases) and L. major (12 cases) using LITSR and L5.8S primers and HaeIII digestion (21). In addition, Doudi et al. used HaeIII and TaqI digestion in isolates from Isfahan and Bam, resulting in two L. major (LmA and LmB) and two L. tropica (LtA and LtB) strains, with digestion patterns similar to ours (140 and 210 bp); however, TaqI digestion generated 150 and 200 bp fragments, indicating a difference between central Iran isolates and isolates from Dasht-e-Azadegan (10). Tashakori et al. determined the genotype heterogeneity in zoonotic cutaneous leishmaniasis endemic areas of the country (Damghan, Dehloran, Dezful, and Kashan) and identified five strains (LmA, LmB, LmC, LmD, and LmE). Their results suggested that LmA is dispersed throughout Iran, especially in the southwestern part. LmC, LmD, and LmE were found only in Damghan and LmB was the exclusive isolate in Kashan (28).
Averting possible mutations, development of drug resistance, and subsequent generation of new strains would be dependent on the genetic persistence of a microorganism in reservoir hosts, thus preventing the impact of harsh environments. Consequently, LmA of L. major with rodent reservoir hosts naturally occurs in areas with L. major dominance. Furthermore, it is suggested that LmA is the most ancient strain with significant adaptive capabilities to conditions of new foci and other genotypes have deduced from LmA. On the contrary, anthroponotic L. tropica does not possess reservoirs; thereby, it is more exposed to hostile milieu (environmental, seasonal, treatments, etc.) and probable mutations in the genome. Hence, the formation of novel strains is more likely to occur in L. tropica than in L. major (8, 10, 29).
Previously, a number of molecular studies on the diagnosis of cutaneous leishmaniasis were done in the Southwestern endemic region, particularly Khuzestan province. In a comparative study by Khademvatan et al. in some parts of the province, 216 skin biopsies from cutaneous leishmaniasis patients were examined using microscopy, culture, and mini-exon PCR; the authors remarked that the molecular technique is the gold standard technique that could diagnose both L. major and L. tropica (11). Another molecular study used real-time PCR and melt-curve analysis by Kinetoplast DNA primers on 102 archived slides, emphasizing L. major as the predominant strain (93.75%), based on our results in the southwest of Iran (30). A nested-PCR approach was also performed on 146 clinical samples in four geographical regions of the province by Maraghi et al., again with L. major as the prevailed species (94.5%), followed by L. tropica (5.5%) (31). In addition, a mini-exon PCR-RFLP study was conducted on 14 clinical cutaneous lesions of suspected patients from a different region of the province; the results identified 12 cases as L. major and only two cases as L. tropica. The authors used EaeI and HaeIII, whereas they did not determine parasite strains, unlike to our study we did (13).
5.1. Conclusions
In total, our results obtained by the PCR-RFLP method showed leishmaniasis, in particular, due to L. major, is predominated in Dasht-e-Azadegan region, Khuzestan province, suggesting the importance of zoonotic cutaneous leishmaniasis in the west and southwest endemic foci particularly with the presence of rodent reservoir hosts in the Khuzestan plain. Hence, future preventive measures should be taken into account. It is also suggested that the species/strains of Leishmania parasites be identified in isolates from rodents and sand fly vectors dwelling the area.