1. Background
Human enteroviruses are members of Picornaviridae family which are divided into four groups consisting of A, B, C and D (1). Hand, Foot and Mouth Disease (HFMD) is a mild disease which is predominant in the Asia-pacific region in infants and children. Enterovirus 71 is an important virus responsible for HFMD which belongs to group A Enteroviruses (2, 3). Enterovirus 71 was detected in 1969 in California for the first time from a child with encephalitis (4). Hand, foot and mouth disease infection usually causes exanthems but neurologic disease such as meningitis, encephalitis and flaccid paralysis also could be seen in patients (5). Different outbreaks of HFMD infection due to Enterovirus 71 have been seen in different parts of the world including Southeast Asia, Europe and Australia (3, 5-12).
Enteroviruses have a single-stranded positive-sense RNA genome which has 4 structural proteins (VP1-VP4) and 7 nonstructural proteins (2A-2C and 3A-3D) (13, 14). It has one large open reading frame (ORF) which produces a large polyprotein (15) 2A and 3C are two proteases in enteroviruses which are very important for virus maturation because they are responsible for the cleavage of viral polyproteins. 2A is responsible for the first cleavage in polyprotein but most of the cleavages are done with 3C protease. The 3C protease is a cysteine protease belonging to the chymotrypsin protease family which is responsible for most of the cleavages on Enterovirus polyprotein (8 out of 10 cleavages on the viral polyprotein). The first cleavage between P1 and P2 region is done by 2A protease but other cleavages, especially in nonstructural regions, are done with 3C protease and it’s precursor 3CD (16).
The following amino acid positions P4, P2, P1, P1’, and P2’ are important for cleavage substrate sequence by 3C. P1 and P1’ positions show the highest conservation in 3C protease, the glutamine or glutamate for P1, and glycine, asparagine or serine for P1’ are very common amino acids in these sites. In addition, the most common residue in position P4 is alanine and in position P2’ is proline. Mostly, 3C detects the cleavage site at the Gln/Gly amino acid pair (16-18). Interferon cytokine is divided into three classes consisting of type I, II and III, in which type I expresses in mammalian cells (19). Theresponse of interferon (IFN) is induced in two levels, IFN production is induced after viral infection and then produced IFN attaches to IFN receptors and induces the IFN signaling pathway.
Viral DNA and RNA or intermediate replicative double-stranded RNA (dsRNA) are recognized as pathogen-associated molecular patterns (PAMP) by pattern recognition receptors (PRR) such as Toll-like receptors (20) and RIG-1 like receptors (RLR) which result in IFN α and β production (21, 22). Produced IFN is released from cells and attaches to IFN receptors on adjacent cells. Two protein kinases including Janus kinase1 (JAK1) and tyrosine kinase 2 (TYK2) are phosphorylated, which leads to activation and phosphorylation of STAT1 and STAT2 proteins and interacts with IRF9 to produce (ISGF)3 complex (interferon-stimulated gene factor 3) , this complex is translocated to the nucleus and induce ISGs (interferon stimulated genes) expression (23). Hung et al. claimed that 3C protease as a main protease of Enteroviruses has a negative effect on the IFN signaling pathway (24), but a recent report revealed that 3C does not have such an effect on IFN signaling pathway (25).
2. Objectives
Based on these differences the aim of this study was the evaluation of Enterovirus 71 3C protease effect on IFN signaling pathway as an important and functional protease in enteroviruses.
3. Methods
3.1. Designing of 3C Plasmids
A plasmid consisting of 3C protease- linker- cleavage site sequence (3C-L-CL) was designed using the 3C gene belonged to Enterovirus 71 strain BrCr. Two kinds of linkers, including a rigid linker (A(EAAAK)n (n = 4)) and a 3C cleavage site (RTATVQGPSLDFE) were placed after the 3C sequence with a combination of 5 restriction sites as follow respectively: XhoI, SacI, SacI, HindIII and SacII. These cleavage sites were added to the start of the 3C sequence, between 3C and linker sequence, between linker and cleavage site sequence and end of the cleavage site sequence respectively. With this structure, we could check the functionality of 3C protease and give the ability of self-cleavage to the 3C protease. The synthesized plasmid (Biomatic, Canada) was subcloned in the pEGFP-N1 vector. The second form of 3C-L-CL plasmid, including 3C-L (linker) was prepared by removing the CL (cleavage site) by enzyme digestion using XhoI and HindIII to show the difference between released 3C and fused 3C to GFP and select one of them to use in our study. After transformation of these plasmids in DH5α with the CaCl2 method, the plasmids were cultured in LB Broth media and extracted using Plasmid DNA Extraction Mega Kit (Qiagen, Germany).
3.2. Rhabdomyosarcoma Cell Transfection with 3C Plasmids
Rhabdomyosarcoma cells were cultured in DMEM growth medium in 24 well plates until 80% confluency. Transfection of RD cells was done by calcium phosphate method using 2X BBS buffer briefly as follows: 100 µL CaCl2 (0.25 M) was added to a 1.5 mL microcentrifuge tube and then 5 µg plasmid was added. The same volume of 2X BBS (calcium phosphate N, N-bis(2-hydroxyethyl)-2-aminoethane sulfonic acid buffered saline) buffer was added to this mix and incubated for 15 to 20 minutes at room temperature. After incubation, whole of the mixture was added to DMEM growth culture media. Twenty-four well plates were kept at a 37°C incubator with 3% CO2 for 6 hours. Afterwards, DMEM media were removed and cells were washed twice with PBS to eliminate plasmids precipitate from the cells and fresh growth medium was added. Cell culture plates were incubated at 37°C with 5% CO2 for one to three days. The RD cells were checked every day under a fluorescent microscope to evaluate the protein expression using observation of GFP reporter.
3.3. RNA Extraction and Real-Time PCR for Evaluation of ISGs Expression
Forty-eight hours after transfection of RD cells by 5 µg 3C-L-CL plasmid in 6 well plates by the calcium phosphate method, cells were treated with 550 IU/mL IFNβ (Sinagen, Iran) for 4 hours. Total RNA was extracted by Tripure solution (Roche /Germany), extracted RNA was treated with DNase to remove the possible trace of DNA and subjected to cDNA synthesis using a cDNA synthesis kit (TAKARA/USA). All cDNA synthesis, transfection, and real-time PCR assays were done in duplicate and intact pEGFP-N1 and untransfected cells used as controls. Each sample was subjected to real-time PCR assay by specific primers for ISGs mRNA (OAS and MxA) and GAPDH as a control. Following primers were used: MxA forward: CAGCACCTGATGGCCTATCA, MxA reverse: ACGTCTGGAGCATGAAGAACTG, OAS1 forward: TCCACCTGCTTCACAGAACTACA, OAS1 reverse: TGGGCTGTGTTGAAATGTGTTT, GAPDH forward: GGTCTCCTCTGACTTCAACA, GAPDH reverse: AGCCAAATTCGTTGTCATAC. A master mix of the PCR reactions, containing 12.5 µL 2× real-time PCR master mix (Qiagen), 0.6 µL of each primer (10 PM), 2 µL template and double distilled water up to 20 µL, was prepared for a PCR program of 40 cycles with the following condition: 95°C for 3 minutes as initial denaturation, 94°C for 10 seconds, 55°C for 10 seconds, 72°C for 10 seconds and 72°C for 2 minutes as final extension.
3.4. Western Blot Analysis for the Evaluation of Interferon Signaling Pathway Proteins
Forty-eight hours after transfection of RD cells, DMEM media was removed and the cells were washed twice with PBS. Then RD cells were lysed on ice with 300 µL RIPA lysis buffer supplemented with protease inhibitor cocktail (Melford, UK). After that plates were incubated for 30 minutes on ice or in the refrigerator. After complete lysis, cells were harvested in a 1.5 mL microcentrifuge and cell clamps were disassociated with sonication three times for 4 to 6 seconds, all the process was done on ice. The lysates were centrifuged for 5 minutes in 4°C with 16000 X g and sample buffer was added. Lysates were incubated at 95°C for 10 minutes and then were kept at -20°C. For further analysis, 50 µL of cell lysate was loaded on 12% polyacrylamide gel and electrophoresed in electrophoresis buffer (25 mM Tris, 192 mM glycine, pH: 8.3) by 120 volts. After electrophoresis, a wet transfer system was prepared for transfering proteins into the nitrocellulose membrane.
Briefly, polyacrylamide gel was put on the nitrocellulose membrane between two pads soaked in transfer buffer (25 mM Tris, 192 mM glycine. 20% methanol, pH: 8.3) to make a sandwich. This sandwich was put in the bio-rad transfer system and the power was set to 100 volts for one hour. After transfer completion, the membrane was blocked with 5% skimmed milk solution with TBST overnight at 4°C. The membrane was washed four times with TBST buffer and specific antibodies added and incubated overnight at 4°C. Three specific antibodies were used in this study, including anti 3C (gene Tex), anti-GFP (Biolegend) and anti-βactin (Santa Cruz) antibodies which were diluted respectively as follows, 1:3000, 1:500 and 1:500. After the first incubation, the membrane was washed four times with TBST and incubated at room temperature with HRP conjugated secondary antibody for two hours. HRP conjugate secondary antibodies (Santa Cruz) dilutions were 1:5000 for 3C, 1:10000 for Bactin and 1:5000 for GFP. ECL substrate was added to the membrane and protein bands were visualized under the ChemiDoc camera (Syne gene, UK).
4. Results
4.1. Transfection of Designed Constructions
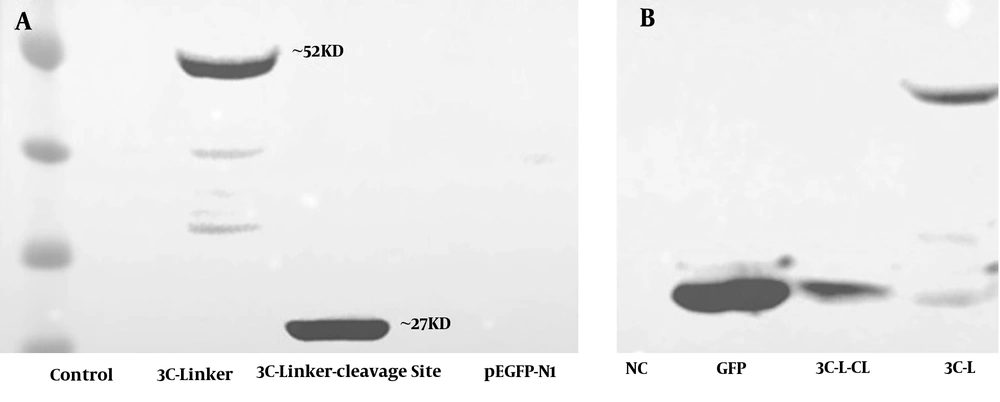
Rhabdomyosarcoma cells were cultured and transfected with 3C-L-CL and 3C-L constructions and cells were checked every day to record GFP expression. After 48 hours, acceptable GFP report was seen in control cells (RD cells transfected with pEGFP-N1). The GFP report from two other designed constructions was very weak compared to the control, Figure 1A - C. Weak GFP expression in two constructions could be for protease activity of expressed 3C. The 3C protease was expressed in both constructions and detected by Western blot analysis, Figures 1D and 2.
A, Western blot results of RD cells transfected with 3C-L-CL plasmid. Negative control: RD cells which did not transfect with any plasmid, 3C-linker: RD cells transfected with a plasmid containing 3C protease and rigid linker, 3C-Linker-cleavage site: RD cells transfected with a full plasmid containing 3C protease, rigid linker, and 3C cleavage site. In 3C-linker plasmid, 3C and GFP have been attached together using rigid linker and create ~52 KD band. In 3C-linker-cleavage site plasmid, 3C cleaves the polyprotein at cleavage site and release itself from the whole polyprotein. B, Western blot analysis of RD cell lysates by anti GFP polyclonal antibody. NC: negative control, GFP: Expression of GFP in RD cells transfected with pEGFP-N1 vector, 3C-L-CL: 3C release itself by autocleavage because the construction has 3C cleavage site (3C-L-CL) and ~27 KD GFP band is observable, 3C-L: 3C-L plasmid does not have 3C cleavage site (3C-L) so 3C remains attached to GFP and create ~52 KD band. The weak GFP report in two plasmids could be the effect of protease activity of 3C on GFP protein.
4.2. Interferon-β Induced Interferon Signaling Pathway Even with 3C Protease Treatment
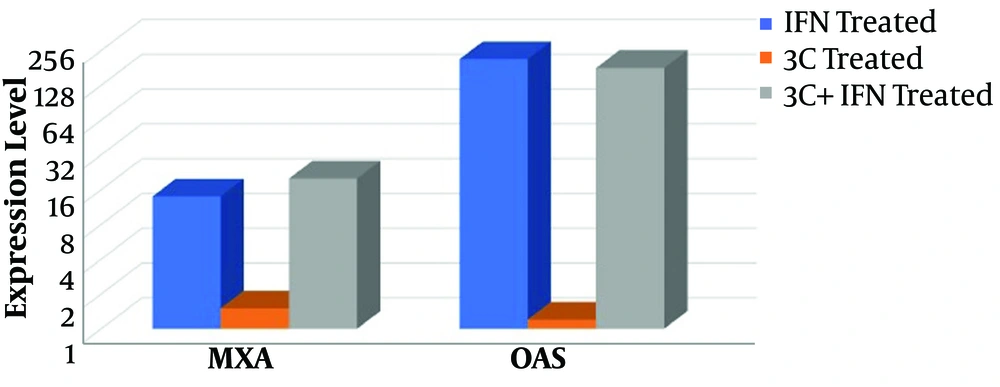
For evaluation of the 3C protease effect on IFN signaling pathway, RD cells were transfected with 3C-L-CL plasmid and treated with IFN-β then the expression level of the MxA and OAS1 genes were screened. The expression level of two ISGs was significantly upregulated by the induction of IFN in control groups and treated group (cells transfected with 3C plasmid). As it is clear in Figure 3 MxA mRNA level increased 14 and 20 fold in RD cells treated with IFN and RD cells treated with 3C+IFN respectively and OAS mRNA level increased 216 and 181 fold in the two mentioned groups. In the OAS group, a little reduction of mRNA expression is observable in the 3C+IFN group but it was not significant. In fact, 3C protease cannot inhibit the upregulation of ISG mRNA induced by IFN and the difference between 3C treated and control group was not significant, P > 0.05, Figure 3.
4.3. 3C Protease Does Not Cleave or Reduce Interferon Pathway Proteins
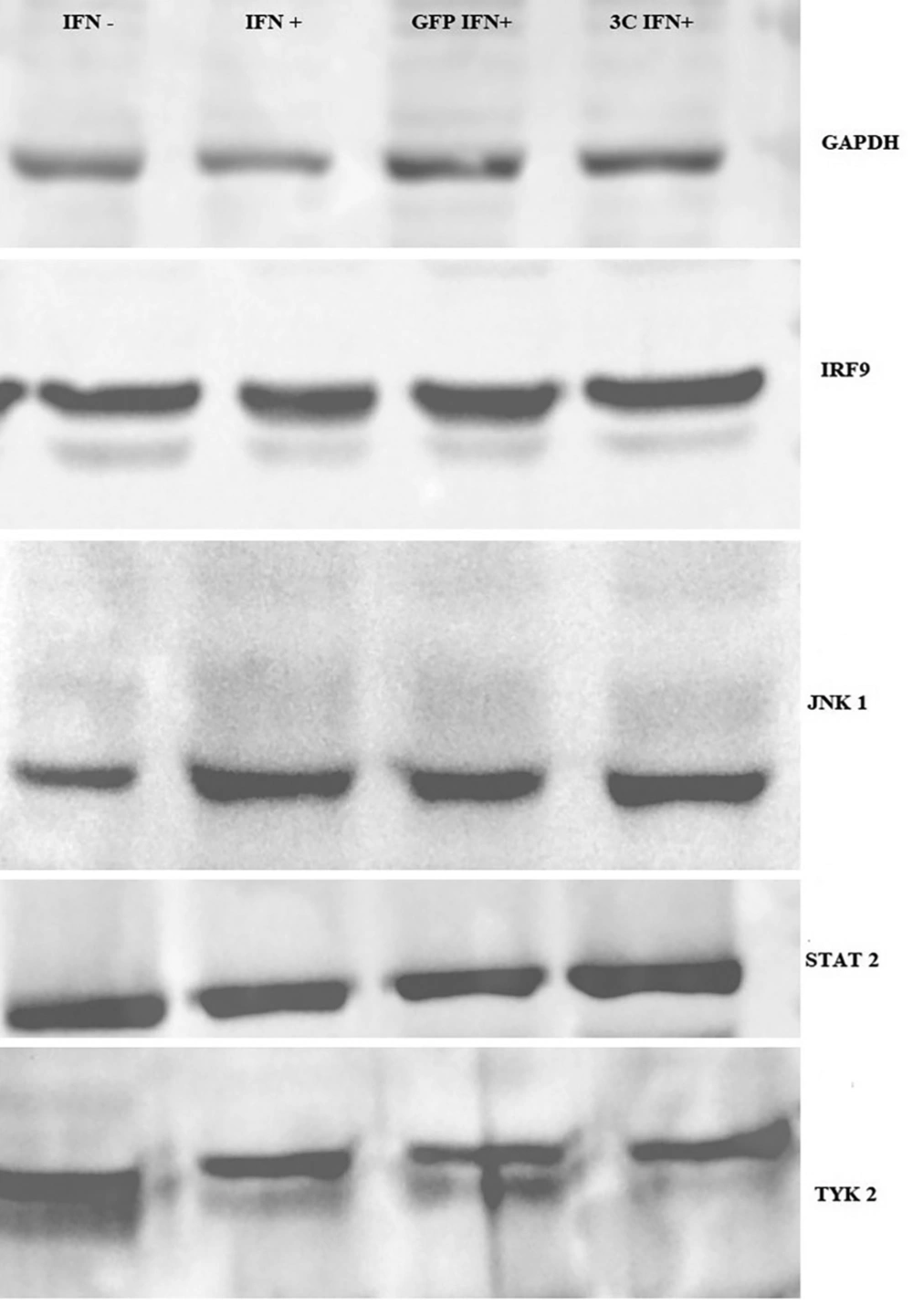
Four proteins of the IFN signaling pathway, including STAT2, TYK2, JNK1, and IRF9 were evaluated for possible cleavage or reduction by 3C protease. Fourty-eight hours after RD cell transfection by a 3C-L-CL plasmid, cells were lysed using RIPA buffer containing a protease inhibitor cocktail and subjected to Western blot analysis. Rabbit polyclonal antibodies (Santa Cruz) of STAT2, TYK2, JNK1, and IRF9 in combination with mouse anti-rabbit HRP conjugate which were used to detect proteins. As it is observed in Figure 4, there is not any significant difference between samples which were transfected with 3C plasmid compared to control groups, including untransfected RD cells and RD cells which were transfected with intact pEGFP-N1 plasmids.
5. Discussion
Enterovirus 71 is one of the most important causes of HFMD and a major concern after poliovirus because both of them cause flaccid paralysis. Enterovirus 71 outbreaks have been seen in different countries, mostly in Asia (26). This virus has some tools to overcome the native immune system response. Poor IFN induction in infected cells was reported in some studies and others indicated that cells should be treated with high dose IFN to protect cells from infection (26, 27). In contrast, in HT-29 cells, Enterovirus 71 induced IFN production better than RD and Hella cells. These findings indicate that different tissues and cells show different interactions to Enterovirus 71 infection. In the IFN production cascade TRIF is reduced significantly in the 12 hours after RD cells infection and usually is not seen in the 36 hours after infection, but the TRIF level does not show any change in HT-29 cells after infection with Enterovirus 71 (27).
Enterovirus 71 overcomes the host native immune system by affecting the IFN signaling cascade, this cascade is induced by IFN and leads to expression of ISGs like OAS1 and MxA. These ISGs have negative results on virus replication and maturation. After IFN attachment to IFNR (interferon receptor), a cascade of proteins like JNK1, TYK2, STAT1, and 2 and IRF9 are activated by phosphorylation which leads to expression of ISGs. The virus can disrupt this pathway and inhibit IFN response (28). Lu et al. infected RD cells with Enterovirus 71 and treated with or without interferon and revealed that 2A protease reduce IFNR1 level and suppress IFN signaling cascade 6 hours after infection (23) but Liu et al. (29) used MRC-5 (human embryonic lung fibroblast) or RD cells infected with Enterovirus 71, and treated with IFN-α2b. This study indicated that Enterovirus 71 inhibits the cellular type I IFN antiviral pathway by downregulating JAK1. In fact, no change in IFNR level is observable after infection and Enterovirus 71 block JAK1 and TYK2 phosphorylation by JAK1 downregulation.
Interestingly, no change in JAK1 mRNA level has been reported; but protein level has been reduced which indicates a disruption in translation stage (29). Liu et al. hypothesized that maybe interaction of more than one viral protein is responsible for this reduction and 2A or 3C do not affect IFN signaling pathway. On the other hand, some studies reported that 3C protease can cleave IRF9, an effective adaptor in the IFN signaling cascade and inhibit ISRE promoter activation (24). Wang et al. though, suggest a new hypothesis for the IFN signaling cascade disruption which is different from previous reports. This study indicates that Enterovirus 71 does not affect IFN signaling cascade by phosphorylation suppression of STAT1 or IFNR1 and JAK1 downregulation, but inhibits their translocation to nucleus by disrupting the interaction between P-STAT1 and KPNA1 using the degradation of KPNA1 (25).
These reports indicate that there is no similar hypothesis about the 3C role in the inhibition of the IFN signaling pathway. Based on these different hypotheses, we evaluate 3C protease effects on IFN signaling pathway as the most important protease of enteroviruses. Our results in this study were in accordance with Wang et al. (25) and Liu et al. (29) reports, our study demonstrates that 3C does not inhibit the IFN signaling pathway. As observable in Figure 3, two ISGs, Mxa and OAS do not affect by 3C protease treatment. There was no difference between ISGs expression in IFN+ and 3C-IFN+ group, in both of them Mxa expression level increased ~20 fold and OAS increased ~181 fold, Figure 3. A little reduction in OAS expression level was observed (181 fold in 3C+IFN group compared to 216 fold in IFN+ group) but was not significant; similar to Wang et al. (25) which reported small but nonsignificant changes in expression level of some ISGs in cells treated with 2A and 3C proteases. Four proteins of IFN signaling cascade were also evaluated by Western blot analysis for observation of possible reduction in expression level or signs of cleavage.
As indicated, there is no difference in translation level and possible cleavage in these proteins between 3C treated and control group. Wang et al. also demonstrated that even phosphorylation of IFN signaling pathway proteins such as STAT1 and 2 and JNK1 which are reported in previous studies were not affected by 3C or 2A protease and using luciferase assay indicated that IFN pathway works normally in the presence of ectopic 2A and 3C, but KPNA1 degradation has the main role in the suppression of the IFN signaling pathway in Enterovirus 71 infection. While KPNA1 is responsible for protein translocation to nucleus, this protein degraded in a caspase 3 dependant manner and neither 2A or 3C have no role in KPNA1 degradation (25). Our result is in accordance with Wang et al., (25) which demonstrates that IFN signaling pathway suppression has no relation with 3C protease activity of Enterovirus 71.
This study revealed that host and viral protein interactions are complex which are affected by different factors such as tissue and cell type, immune system situation of host and different kinds of host. Maybe more evaluation of other cells leads to different and new results about this interaction between Enterovirus 71 proteases and IFN signaling cascade. It will also be interesting to focus on the role of other viral proteins on IFN pathway inhibition. These findings indicate that viral and host protein interaction is multifactorial and needs more research to gain a wide and better view about viral pathogenesis and host response.
5.1. Conclusions
3C protease does not reduce ISG expression level and IFN pathway protein such as STAT2, IRF9, TYK2 and JNK1 and could not suppress IFN signaling pathway alone.