1. Background
Hepatitis C virus (HCV) is a human flavivirus, a positive sense, single-stranded RNA virus that causes both acute and chronic hepatitis. This viral agent is a frequent public health challenge globally (1, 2). Overall, 185 million people (3% of the world population) are suffering from HCV infection; however, this rate has been fewer than 2% in Iran (3). The main HCV-infected population in Iran and other parts of the world are injecting drug users (IDUs) (4). Many patients infected with HCV neglect to clear the infection biologically, resulting in a chronic phase of the infection (5) that may progress toward liver cirrhosis and also hepatocellular carcinoma (6). No vaccine has yet developed to immunize individuals against HCV infection. A combination of Interferon-α (IFN-α) plus Ribavirin (RBV) has been considered as the standard HCV therapy during the last decades. Nevertheless, this combination did not show complete effectiveness in some patients, especially in patients with HCV genotype 1 (7). Although the introduction of direct-acting antivirals in recent years has created new hopes in the effective treatment of chronic HCV, the therapeutic regime is still an IFN-ribavirin combined regimen in low-income regions (8, 9). On the other hand, the treatment of HCV, as an infectious disease, provides a suitable model that reveals many aspects of the interaction between interferon and host cellular mechanisms.
Autophagy is a fundamental event in eukaryotic cells, such as hepatocytes, in which double-membrane structures named autophagosome can be formed. Theses compartments engulf long-lived cytoplasmic macromolecules and injured organelles and also conduct them to lysosomes for decay and/or recycling (10, 11). It is a constitutive process that usually occurs at the basal level; however, it can be increased due to extracellular or intracellular stresses and also factors such as endoplasmic reticulum stress, starvation, viral infection, and growth factor depletion (12, 13). As intracellular parasites, viruses interact with and subvert the autophagy pathways during the infection cycle. In the infection cycle of some viruses, such as human herpes simplex virus (HSV), Sindbis, and vesicular stomatitis virus (VSV), autophagy play as a cell-intrinsic antiviral immune defense and other viruses, like HSV and Epstein–Barr virus, autophagy is involved in adaptive and innate immunity.
Kaposi’s sarcoma-associated herpesvirus (KSHV), HSV, Cytomegalovirus, Epstein–Barr virus, human immunodeficiency virus, and some other viruses suppress the induced autophagy mechanisms in infected host cells through different strategies. Beclin-1 is an ortholog of the Atg6 yeasts with a critical role in the autophagy process. BH-3 domain of this protein accompanies with several cofactors that can interact with Bcl-2 and Bcl-XL and inhibit them (14, 15). Autophagosome formation in dengue virus, poliovirus, influenza virus A, and coxsackievirus B3 virus infections is associated with improved viral replication and also an increase in viral yield (16-18). Different studies have demonstrated autophagy induction following HCV infection in host cells, which is independent of HCV genotypes.
Notably, both full-length and subgenomic HCV RNA, as well as HCV virions, can induce autophagic vacuoles (10, 11, 19, 20). It has also been proved that the knockdown of autophagy prevents the generation of infectious virus particles. Shrivastava et al. have eventually demonstrated that the disruption of the autophagy process in HCV-infected hepatocytes stimulates the interferon signaling pathway, leading to an improvement in the innate immune response (21). Wang and Ou reported that both HCV structural and non-structural proteins potentially regulate the autophagic pathway. For example, HCV core is a known protein that can induce endoplasmic reticulum stress, and subsequently activate the autophagic pathway (22).
2. Objectives
This study aimed at assessing the expression level of Beclin 1 and IFN-α in chronic HCV-infected individuals using real-time PCR assay, as a fast and high-throughput detection test. Two groups of IFN-ribavirin responders and non-responders were evaluated to compare the effect of autophagy on the antiviral mechanism of IFN-α.
3. Methods
3.1. Patients
This research was conducted on patients (age range, 17 - 64 years) with confirmed chronic HCV referred to the Professor Alborzi Clinical Microbiology Research Center, Namazi Hospital, Shiraz, Iran, during 2015 - 2017. In this study, a total of 40 samples from peripheral blood mononuclear cells (PBMCs) were evaluated. Twenty samples were collected from the patients with chronic HCV, who were non-responders to treatment, and 20 samples were obtained from the responders to treatment with peg- IFN-ribavirin following up one year after treatment. All samples were taken in the pre-treatment period. Notably, patients infected with HCV genotype 1 were selected according to the available profiles. Also, ten samples of healthy people who had no infectious disease for two weeks before sampling were collected as the reference samples.
3.2. PBMC Isolation
Five milliliters of the whole blood was diluted with PBS and then centrifuged on top of 3 mL of ficoll in 400 g for 30 minutes in 18 to 24ºC. The PBMC layer was separated using a sampler and washed with the same volume of PBS.
3.3. RNA Extraction and cDNA Synthesis
RNA extraction was done using a High pure RNA isolation kit (Roche, Germany), according to the Manufacturer’s guideline. The cDNA synthesis was carried out using the QuantiTect reverse transcription kit (Qiagen, Germany), according to the manufacturer's protocol. Briefly, the purified RNA was incubated in gDNA Wipeout Buffer at 42°C for 2 minutes to eliminate genomic DNA. One microgram of the RNA sample was added to the reverse-transcription master mix containing random and oligo-(dT) primers. The mixture was incubated at 42°C for 15 minutes and immediately placed at 95°C for 3 minutes to inactivate reverse transcriptase.
3.4. Real-Time PCR Assay
The used primers were designed using Lasergene 7 software. The related sequences to human IFN-α, Beclin 1, and GAPDH genes were downloaded from nucleotide data bank in FASTA format. The sequences were aligned for each gene, and the conserved areas were characterized for designing the primer. The primers were designed from separate exons or connection areas of the exons. The designed primers were evaluated for specificity in recognizing the target gene in the NCBI Primer Blast site. Table 1 represents the primer sequences designed by Lasergene with the length of the segment and the length that the corresponding pair primers are proliferated. Real-time PCR reactions were performed using the SYBR Green reaction kit according to the manufacturer’s instructions (Takara, Japan) in ABI 7500 system. Also, cDNA (2 μL each) was diluted to a volume of 20 μL with the PCR mix containing a final primer concentration of 0.2 ρmol. The samples were tested in triplicate, and the expression level of each target gene was analyzed by the 2-ΔΔCT method. The ΔCT values were normalized to GAPDH for each triplicate set.
| Gene, Premier | Sequence | Length, bp |
|---|---|---|
| IFN-α | ||
| Forward | CTCAAGCCATCTGTGTCCTCC | 171 |
| Reverse | CTACCACCCCCACCTCCTGT | 171 |
| Beclin 1 | ||
| Forward | CGCTGAGGGATGGAAGGGTCTAAG | 163 |
| Reverse | CCTGGGCTGTGGTAAGTAATGGAG | 163 |
| GAPDH | ||
| Forward | ACCTGACCTGCCGTCTAGAAA | 68 |
| Reverse | CCTGCTTCACCACCTTCTTGAT | 68 |
3.5. Data Analysis
The collected data were statistically analyzed using SPSS or Graph pad software. The correlation between Beclin and IFN-αin, each group, were analyzed by Pearson's correlation coefficient.
4. Results
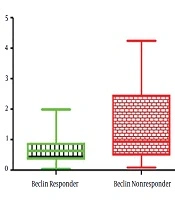
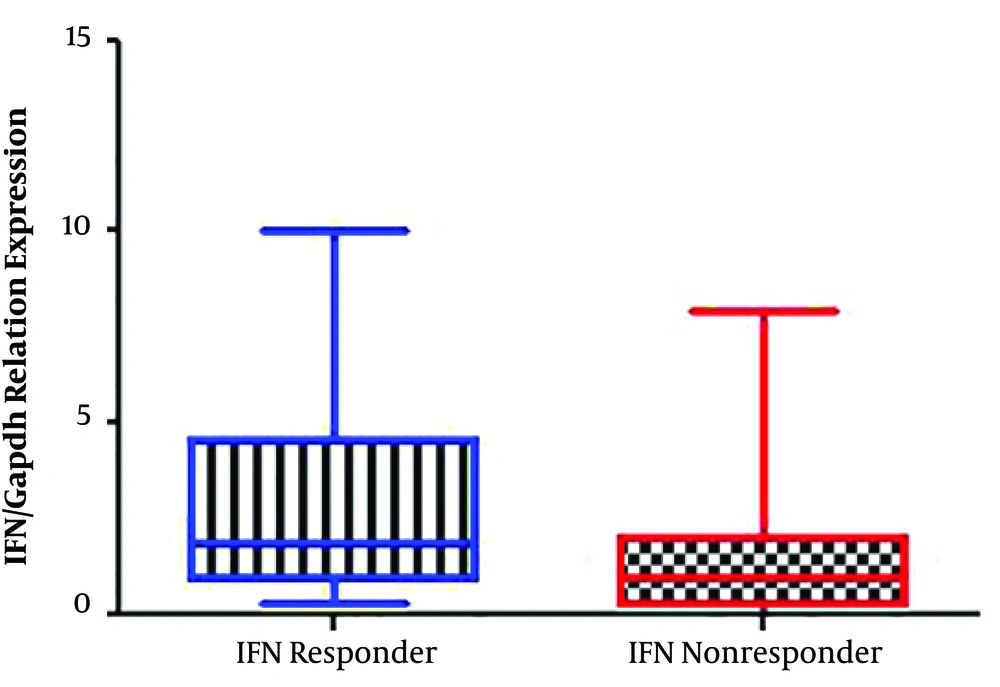
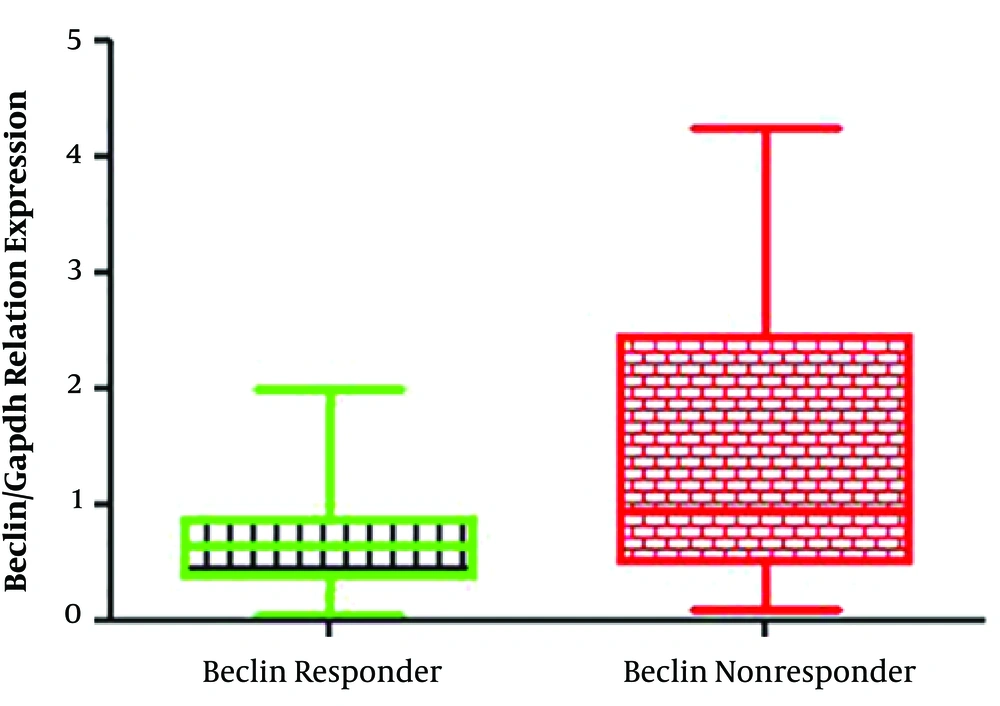
In the present study, 40 patients, including 20 non-responders and 20 responders to the peg-IFN-ribavirin treatment with the mean age of 44.18 ± 12.02 years, were recruited. The pre-treatment HCV viral load found to be 4.2 × 106 ± 8.8 × 106 in responder group, which was significantly lower than the non-responder group with a viral load of 1.1 × 107 ± 9.1 × 106 (P < 0.02). The expression rate of IFN in the responder group was significantly higher compared with the non-responders (P < 0.02) (Figure 1), whereas the expression level of the Beclin 1 gene was significantly lower in responders (P < 0.02) (Figure 2). Furthermore, a negative correlation was found between Beclin 1 and the IFN-αexpression level in the non-responder group (r = -0.56; P = 0.05).
The expression level of the IFN gene in IFN-ribavirin responders and non-responders with HCV infection. As shown in the figure, the expression level of the IFN gene was significantly higher in responders than non-responders. Gene expression level was determined in triplicate per sample using the 2-ΔΔCt method.
The expression level of the Beclin 1 gene in IFN-ribavirin responders and non-responders with HCV infection. The expression level of the Beclin 1 gene in responders was significantly lower compared with non-responders (P < 0.02). The relative changes in IFN gene expression were analyzed using the 2-ΔΔCt method. All experiments were performed in triplicate.
5. Discussion
Peg-IFN/ribavirin treatment has been known as a standard treatment for cases with chronic HCV. Various studies have shown that factors, such as genetics, obesity, age, gender, the genotype of HCV, and the rate of viral load, are useful in response to treatment (23, 24). Different studies have been conducted to clarify the role of autophagy in the process of chronic HCV infection, and several investigations have addressed the role of autophagy in HCV infection, the mutual role of HCV infections and autophagy, as well as the role of autophagy in IFN production. However, no study has yet been done on the role of autophagy in response or non-response to IFN treatment in chronic HCV infections (10). Although new direct-acting antivirals (DAAs) have recently replaced with IFN therapy, the present study on the expression level of Beclin 1 and IFN-αin both responder and non-responder groups demonstrated the importance of autophagy activity as a host factor in IFN production, as well as the outcome of IFN therapy that can be generalized to different diseases.
The results showed that the expression rate of the Beclin 1 gene was significantly lower in the responder group compared with non-responders. Beclin 1 is an essential compartment in the formation of autophagosome and initiating and developing autophagy (25). This protein reacts with other proteins, such as kinase complexes, called Vps34, and other proteins, like Vps15 and Atg14L (26). Shirvastava et al. showed that the expression rate of Beclin 1 in hepatocytes increases in primary stages of infection with HCV. They also evaluated the effect of various proteins of HCV on the promoter area of the Beclin 1 gene, and it was found that NS5A protein plays an essential role in increasing the expression of this protein (21). Various studies were conducted on the effect of HCV infection on the autophagy process, which revealed that the virus stimulates autophagy (27) through an increase in the expression level of Beclin 1. It has demonstrated that HCV develops autophagy incompletely via the response to mal-folded protein (11). It has reported that the virus can disrupt autophagosome maturation and its fusion with lysosomal vesicles and utilizes these structures for its proliferation (11) to prevent viral destruction via the autophagy pathway (28). Other studies conducted on the effect of autophagy in facilitating the HCV proliferation process have shown that this virus benefits from autophagy-related proteins, such as Beclin 1, Atg5, Atg4B, and Atg12 to facilitate proliferation. Therefore, the virus reinforces its proliferation and consequently, its infectivity (10, 28).
Various in vitro and also in vivo studies have revealed that HCV can induce autophagy through different mechanisms. Also, based on the results of in vitro studies, autophagy was induced in various cell lines that were infected with HCV (29-32). In the present study, PBMCs of HCV-infected individuals also showed a high level of Beclin 1expression, which was significantly higher in the non-responder group with a higher viral load. This suggests that more HCV activity leads to increased Beclin 1 expression in blood cells, which can be considered as a factor for a decrease in IFN induction. Both IFN I and II stimulate a similar signaling pathway and provide conditions in the cell that leads to interference with virus replication. The virus also interferes with the IFN signaling pathway using its proteins. NS3 protein has protease activity on its N-terminal, as well as RNA-helicase and NTPase activity on its C-terminal. This protein needs NS4A protein to perform its protease activity. The complex of these two proteins acts as an anti-viral antagonist by inhibiting the activation of transcription of the factors, such as IRF3. Also, this complex prevents the transformation of an intracellular signal resulting from an inherent immunity factor called TLR-3, which also activates IRF3 by inhibiting cell TRIF.
Inactivation of the protease activity of NS3 has been reported unable to restore the promotor activity of IFN B, suggesting an alternative pathway for innate immunity inactivation. It has indicated that the autophagy induced by HCV infection represses the innate immune response through several autophagy molecules that negatively regulate IFN activation by suppression of IFNB promoter activation and reduction of the IFNB mRNA level. Also, the knockdown of autophagy enhances the production of IFNs and decreases the rate of HCV replication, which is consistent with the results of the present study. Our results showed that the expression rate of the Beclin 1 gene in the non-responder group was significantly higher than the responder group, which negatively correlates with the level of IFN expression. This can be considered as a reducing force for IFN-α expression, enhancing the virus replication and non-response to treatment.
5.1. Conclusions
Based on the obtained results, there was a significant difference between the expression of Beclin 1 and IFN-α in PBMCs of the patients infected with chronic HCV who have responded to the treatment with IFN -ribavirin and non-responder patients. Considering the negative correlation between these two factors in non-responders, the enhanced rate of Beclin 1 can be regarded as a potential factor for IFN decline and its effects on virus elimination. Besides, the negative correlation between other genetic and physiological features of the host can be considered as an influential factor in the disability of innate immunity to the virus elimination and non-response to treatment. It can be suggested that the modulation of autophagy may alter the outcome of IFN therapy and also enhance the effect of IFN in virus-infected cells; however, more studies are needed to confirm these results.