1. Background
The World Health Organization (WHO) has recognized the antimicrobial resistance (AMR) as a major global health problem in recent decades (1). Klebsiella pneumoniae, a Gram-negative rod, belongs to the Enterobacteriaceae family. It is an opportunistic pathogen, causing septicemia, pneumonia, urinary tract infections, meningitis, diarrhea, and soft tissue infections. The rising multi-drug resistance (MDR) of K. pneumoniae isolates has led to limited treatment options for antibiotic therapy and infection control (2). Carbapenems are a major class of β-lactam antibiotics for the treatment of serious infections in Gram-negative bacteria. Carbapenemase-producing Enterobacteriaceae (CPE) in hospitalized patients has been a major concern for more than a decade (3). Carbapenemases are a member of molecular classes A, B, and D β-lactamases, that could hydrolyze β-lactam antibiotics (4).
A functional group 2f in Class A carbapenemases have appeared sporadically in clinical isolates for three decades. They founded in Enterobacter cloacae, Serratia marcescens, and Klebsiella spp. (5). The NMC/IMI, SME, KPC and GES enzymes exist in four major families of class A serine carbapenemases. An active-site serine requires at position 70 for a hydrolytic mechanism in the Ambler numbering system for class A β-lactamases. The KPC and GES have two characteristics that separate from the other functional group 2f enzymes (6). First, these enzymes are found on transferable plasmids; they can be transferred from one bacterium to another, from one person to another, and from one country to another (2), second, KPC and GES are clinically important and active against many β-lactam antibiotics include the cephalosporins and carbapenems. Carbapenemases enzymes destroy the antibiotics before they have a chance to have an effect. Due to the potential for transmission of these genes, recognition of KPC-producing organisms is essential for controlling their prevalent in nosocomial and long-term-care settings (7).
2. Objectives
The objective of our study was the detection of blaKPC and blaGES carbapenemases, evaluation of expression level of these genes in the presence and absence of β-lactam antibiotic, and determination of antibiotic resistance patterns among K. pneumoniae isolated at Firoozgar Hospital.
3. Methods
3.1. Study Period, Sample Collection and Bacterial Isolation
This cross-sectional study was conducted in Firoozgar Hospital, Tehran, Iran. We collected K. pneumoniae strains from clinical samples of the patients who were admitted to different wards between March 2018 and December 2018. The K. pneumoniae colonies were identified via standard biochemical tests including indole, motility, citrate, urease, lactose fermentation, lysine decarboxylase, and MR-VP (8).
3.2. Antimicrobial Susceptibility Testing, Modified Hodge Test and E-Test for Imipenem
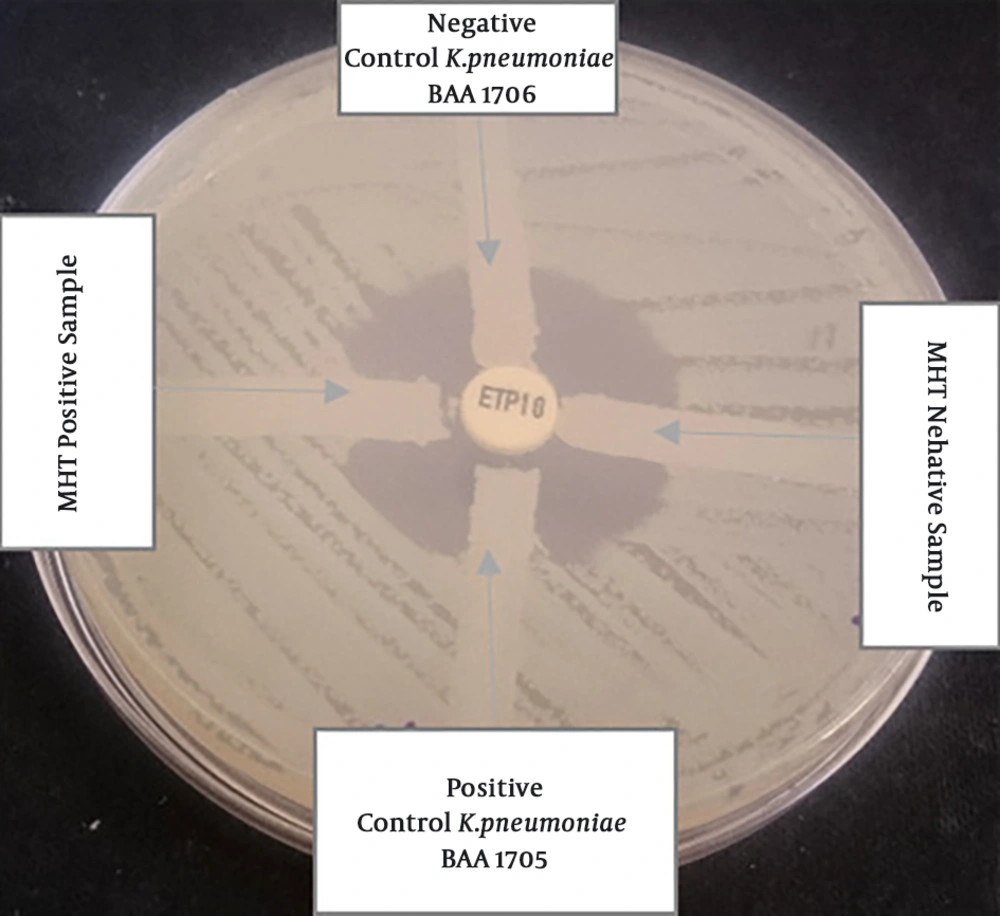
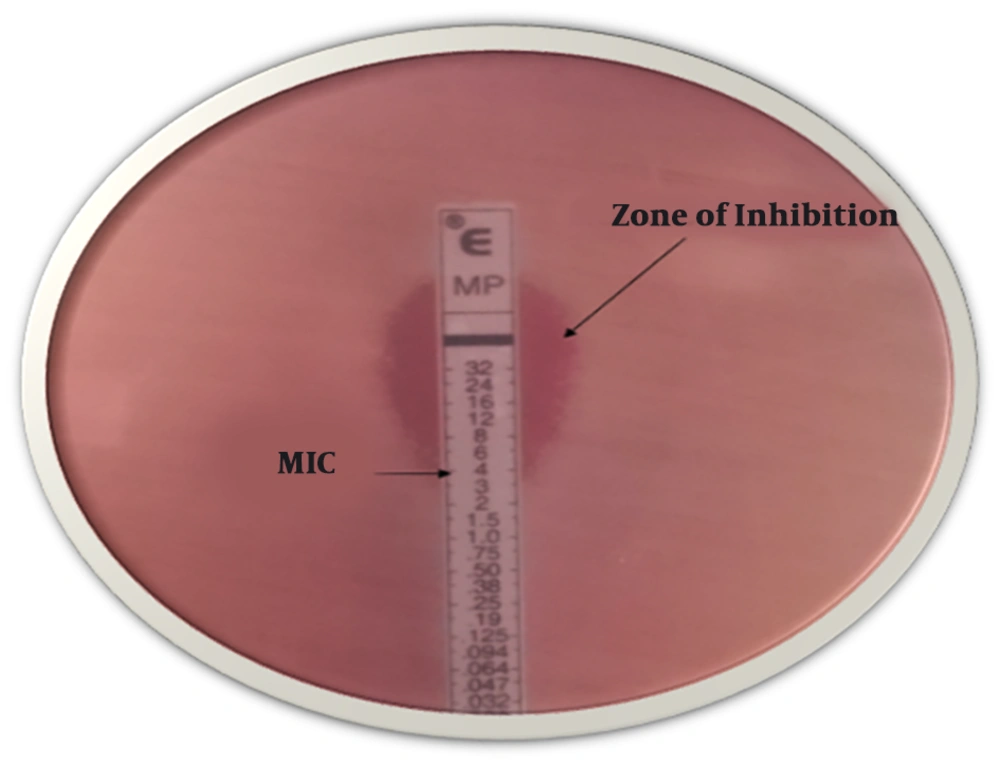
Antibiotic disks (MAST Company, UK) were shown in Table 1, used to perform susceptibility tests via the disk diffusion method, based on the Clinical Laboratory Standard Institute guidelines (CLSI 2016) (9) on the Mueller-Hinton agar plate. Klebsiella pneumoniae ATCC 13883 was used as a control strain (8). Carbapenem-resistant strains were exposed for KPC-producing by the Modified Hodge Test (MHT) based on CLSI guidelines 2016, using Escherichia coli ATCC 25922 and the ertapenem disk (Mast, UK). Positive control (Klebsiella pneumoniae ATCC 1705) and negative control (Klebsiella pneumoniae ATCC 1706) are streaked on the same plate (10). The last test of resistance to carbapenems was determined minimum inhibitory concentration (MIC) for imipenem by using E-test. Results were categorized as sensitive (S), intermediate (I) and resistant (R) as per CLSI guidelines 2016 (S ≤ 1, I 2, R ≥ 4) (11).
| Antibiotic Name | Resistant (R) | Intermediate (I) | Sensitive (S) |
|---|---|---|---|
| Imipenem (10µg) | 100 (55.2) | 15 (8.28) | 66 (36.5) |
| Meropenem (10µg) | 102 (56.3) | 10 (5.24) | 69 (38.2) |
| Ertapenem (10µg) | 102 (56.3) | 14 (7.7) | 65 (36) |
| Doripenem (10µg) | 105 (58) | 10 (5.24) | 66 (37) |
| Cefotaxime (30μg) | 150 (83.5) | 2 (1.1) | 29 (16.02) |
| Ceftazidime (30μg) | 145 (80.1) | 5 (2.76) | 31 (17.12) |
| Cefepime (30μg) | 152 (83.9) | 3 (1.65) | 26 (14.36) |
| Cefoxitin (30μg) | 121 (66.8) | 14 (7.73) | 46 (25.41) |
| Ceftriaxone (30μg) | 148 (81.7) | 4 (2.2) | 29 (16.02) |
| Gentamicin (10µg) | 150 (82.7) | 3 (1.65) | 28 (15.47) |
| Piperacillin (100μg) | 148 (81.7) | 4 (2.2) | 29 (16.02) |
| Aztreonam (30μg) | 108 (59.6) | 13 (7.18) | 60 (33.14) |
a Values are presented as No. (%).
3.3. Molecular Assays
3.3.1. Isolation of Total RNA and cDNA Synthesis
After inoculating the strains from glycerol stocks in nutrient agar (2 mL), they were grown at 37ºC overnight. Next, the strains were sub-cultured in the nutrient medium (5 mL) and grown to the mid-exponential phase (OD600 = 1.5 - 2.0). Afterward, isolation of total RNA was performed based on the manufacturer’s guidelines. DNase treatment was applied to remove the residual DNA with 20 U of RQ1 DNase I (Sinaclon, Iran). A reaction mixture, consisting of 5 µg of RNA, was incubated in DEPC-treated water. Next, cDNA Synthesis was performed based on the manufacturer’s guidelines (Pars Tous, Iran) and the collected cDNA was stored at -20ºC until further analysis.
3.3.2. RT-PCR Technique
Molecular identification of blaKPC and blaGES genes in K. pneumoniae by reverse transcription-polymerase chain reaction (RT-PCR) (12) was applied to evaluate isolates with positive results on the MHT. Specific primer pairs, targeting blaKPC and blaGES genes, were designed by IDT software and synthesized by Macrogen Company (Korea) (Table 2).
| Primer | Sequences | RT-PCR (Step 1) | Real-Time PCR (Step 2) | ||
|---|---|---|---|---|---|
| Conditions | Volume Reactions | Conditions | Volume Reactions | ||
| blaKPC | F- CGGCAGCAGTTTGTTGATTG | 1 cycle: 25ºC - 10 min; 47 ºC - 60 min; 70 ºC - 10 min | Total RNA 5 mL, Random hexamer 2 mL, H2O up to 10 mL | 1 cycle: 95ºC - 10 min, 40 cycle: 95ºC - 30 s; 58ºC - 30 s; 72 ºC - 45 s, 1 cycle: 95 ºC - 30 m | Real Q Plus 2x Master Mix, green: 12.5 μL, Primer F + R Primer: 2 μL, Template DNA: 5 μL, Nuclease-free water: 5.5μL, Total reaction volume:25 μL |
| R- CAGACGACGGCATAGTCATTT | |||||
| blaGES | F- GACTCTGTGAGTCGGCAAGA | ||||
| R-AGCAATAGGCGTAGTTGTATC TC | |||||
| rpo B | F- AACCCGCTGTCTGAGATTAC | ||||
| R-GGCGTTTCGATCGGACATA | |||||
3.3.3. Real-Time PCR Assay
The real-time PCR was used to determine the role of blaKPC and blaGES genes in resistance to carbapenemases, in the presence and absence of imipenem, in 36 antibiotic-resistant clinical strains containing two genes. Quantitative PCR (qPCR) was also performed according to the manufacturer’s instructions. SYBR Green and 2X RT-PCR Master Mix Green were used in a No-Rox kit (Ampliqon, Denmark) and then, an RT-PCR detection system (Corbett, Australia) was used. Based on the melt curve analysis after 40 cycles, the presence of RT-PCR products was examined. The reference housekeeping gene was rpoB gene (Table 2). The Rest software was used to calculate the gene expression, design of the graphs and the critical threshold cycle (CT). Then, the expression analysis was performed by relative measurement of mRNA expression in comparison with Klebsiella pneumoniae ATCC 13883 strain. That way, the comparative Ct (ΔΔCt) method was applied to determine Ct values; fold differences were measured as 2-ΔΔCt. We explained adaptive responses of K. pneumoniae to imipenem-induced stress and investigated these genes that could be involved in resistance to stressful environments. Hene, the strains of resistance to carbapenemases divided into two groups: one group was strains that to exposure 2 mg.L-1 of imipenem and other group was strains without induction antibiotic (13).
3.4. Statistical Analysis
To analyze the obtained data, SPSS version 23.0 (SPSS, Chicago, IL), using a t-test and Pearson’s chi-square test was employed to interpret correlation between parameters, for data analysis were performed of descriptive statistics (frequency, percentage, mean); the level of significance in the current study was < 0.05.
4. Results
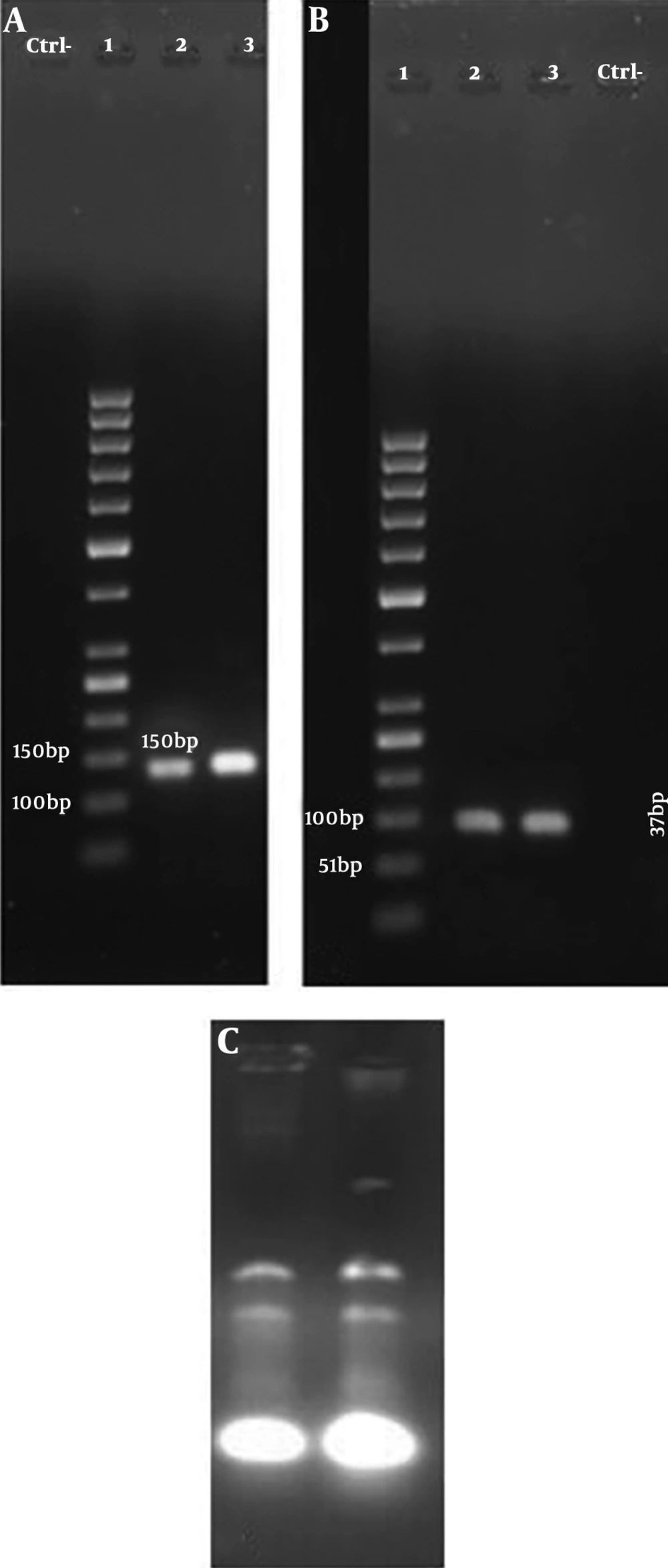
One hundred and eighty-one K. pneumoniae strains were collected from clinical samples such as aspirate (n = 65), sputum (n = 40), urine (n = 35), blood (n = 26) and other clinical samples (n = 15). Among the entire samples, the median patient age was 57.11 years old, 51.38% (n = 93) and 48.61% (n = 88) of isolates were from males and female patients, respectively. Cefepime and imipenem showed the highest and lowest rates of resistance 83.9% and 55.2%, respectively. Klebsiella pneumoniae exhibited the highest susceptibility to meropenem (38.12%) and the lowest susceptibility to cefepime (14.36%). All the samples were confirmed as multidrug-resistant (MDR) strains (Table 1). One-hundred strains were positive as KPC-producing in MHT (Figure 1). 100 strains were exhibited in resistance to imipenem. The MICs ranged from 4 μg/mL to 6 μg/mL (MIC ≥ 4 μg/mL) (Figure 2). The findings of RT-PCR showed that 51% (n = 51) of strains were blaKPC positive, and 49% (n = 49) were blaGES positive; also, 36% (n = 36) of the strains carried both genes (Table 3, Figure 3). There is sufficient evidence for a relationship between resistance to antibiotics and the presence of some genes. We observed the overexpression of these genes when induced by 2 mg.L-1 of imipenem. That caused 1.04- and 12.21-fold increase in the expression of blaKPC and blaGES genes, respectively. That was showed significantly higher than those lacking this antibiotic.
| Gene (s) | Source of Sample | |||||
|---|---|---|---|---|---|---|
| Blood | Sputum | Aspirate | Urine | Other | Total | |
| blaKPC | 7 | 11 | 19 | 9 | 5 | 51 |
| blaGES | 7 | 11 | 16 | 10 | 5 | 49 |
| blaKPC and blaGES | 5 | 10 | 12 | 6 | 3 | 36 |
A, gel electrophoresis shows the PCR product of blaKPC: Lane 1. Gene ruler 50 bp DNA Ladder, Lane 2. PCR product of blaKPC, Lane 3. Ctrl+ (Klebsiella pneumoniae ATCC 13883 strain). B, gel electrophoresis shows PCR product of blaGES: Lane 1. Gene ruler 25 bp DNA Ladder, Lane 2. Ctrl+ (K. pneumoniae ATCC 13883 strain), Lane 3. PCR product of blaGES. C, RNA extracted from K. pneumoniae
5. Discussion
The prevalence of KPC-producing K. pneumoniae in hospitals is associated with increased mortality (14). Carbapenems are often applied as first-line treatment for drug-resistant pathogens. while the increasing frequency of KPC-producing organisms has decreased the efficacy of these antibiotics (15). This study is the first report of the expression of blaGES and blaKPC genes from K. pneumoniae strains of Firoozgar Hospital, Tehran, Iran. Our findings revealed the high prevalence of KPC and GES genes, encoding carbapenem resistance among K. pneumoniae isolates. The gene expression level of blaKPC and blaGES was determined by real-time-PCR and the relationship between higher expression of mention genes and drug resistance was investigated for carbapenem-resistant K. pneumoniae (CRKP) strains.
In this work, 56.45% of the K. pneumoniae strains were resistant to carbapenems. Based on the antibiotic susceptibility tests, the highest resistance to the antibiotic were related to cefepime (83.9%), cefotaxime (83.5%), whereas, the lowest resistance was attributed to imipenem 55.2%, meropenem 56.3%, ertapenem 56.3% and doripenem 58%; that is agreed with the result of Bina et al. (8). MHT was applied as a suitable method for evaluating the production of carbapenemase in the present study. In the MHT, 55.2% (100 of 181) strains were positive for KPC that was consistent with Cury et al. (MHT 71% positive) (11). In three studies, 80.5% (33 of 41), 84% (32 of 38) and 12.3% (30 of 244) strains were positive for KPC-producing organisms’ strains showed the production of carbapenemase (8, 16, 17). Our study showed that the resistant isolates to imipenem by E-test were 100 out of 181 (55.2%), which is in agreement with the results of Gupta et al. They were reported carbapenem resistance by E-test in 10.8% of the isolates (18). Actually, as the last test of resistance to carbapenems confirmed the results of the MHT test.
Detection of KPC carbapenemases by susceptibility testing is not an authentic method. Due to the heterogeneous expression of -lactam resistance by multiple determinants such as AmpC and CTX-M β-lactamase. The method certified by the CLSI is MHT and this method is accepted as a specific and sensitive method for the detection of carbapenemase. However, cannot be used as a confirmatory test for KPC-producing isolates. Also, maybe falsely recognized as carbapenem susceptible, resulting in the inappropriate selection of therapy. The current methods of laboratory diagnosis need to be improved to detect KPC resistance (19). The RT-PCR method provided a convenient molecular tool for the detection and expression of blaKPC and blaGES genes, providing a precaution for the actual outbreak. We showed that 51% of K. pneumoniae isolates which were resistant to carbapenem were positive for the blaKPC gene, which is in line with previous studies from USA (20) and China (21), confirming the presence of blaKPC gene by PCR. Whereas, other studies were negative for the blaKPC gene that had the contrast with our study. This opposition can be related to active drug efflux pump and porin alterations (8, 22). Also, we observed 49 K. pneumoniae strains containing blaGES gene which these results are consistent with Poirel et al. and Picao et al. (23, 24).
With this detail, the molecular detection of carbapenemase genes by PCR has been confirmed as the gold standard. Although the genotypic method is a time-consuming process for a clinical microbiology laboratory because that commonly needs isolates to be referred to reference laboratories for detection (25). Several PCR-based methods have been presented including real-time PCR assays and electrospray ionization mass spectrometry. The real-time PCR method is able to detect rapid and affordable to detected carbapenemases genes, identify variants and measurement expression of these genes (26). In our study, all of the K. pneumoniae strains were MDR, hence real-time-PCR revealed a high expression of blaKPC and blaGES at MDR strains. Moreover, the results of real-time-PCR showed a two-fold increase in the expression of blaKPC and blaGES genes in the presence of 2 mg. L-1 of imipenem. These results are consistent with a four-to eight-fold increase in MexX and mexY gene expression of Pseudomonas aeruginosa in the presence of tetracycline (2 mg/L) (27). Moreover, the gene expression results are in agreement with the study by Dhabaan et al., who showed the overexpression of 12 pilus genes in resistant Acinetobacter baumannii isolates by three folds when treated with a sub-MIC of imipenem (28).
Based on the findings, most of the broad-spectrum antibiotics did not remove carbapenem-resistant K. pneumoniae and the mortality rate of patients is a significant concern. Real-time PCR assay can reduce the turnaround time for infection control measures and the detection of carbapenemase-producing organisms. Overall, the proposed method in this study can be considered an improvement to these methods. We hope that our findings can be applied for the implementation of an effective systematic strategy to manage infectious diseases and prevent the diffusion and dispersion of KPC-producing K. pneumoniae in Iran.
5.1. Conclusions
Due to the high resistance of K. pneumoniae isolates to common antibiotics, the identification of carbapenemase-producing isolates is essential for antibiotic therapy. Also, revisiting the antibiotic therapy protocols for the prevention and control of the spread of resistant bacteria is an effective strategy.