1. Background
The term “leishmaniasis” encompasses different clinical manifestations including cutaneous, visceral, and mucosal forms, which are resulted from the intracellular infection of immune cells by several species of Leishmania parasite. Leishmaniasis is a health problem in most parts of the world and is endemic in more than 88 countries worldwide. The disease mainly affects the tropical and subtropical countries, but 90% of the reported cases are related to Iran, Afghanistan, Brazil, Saudi Arabia, Syria, and Peru (1). In Iran, the disease is distributed in more than half of the provinces and currently, it is estimated that about 20,000 new cases of leishmaniasis are reported annually (2).
In this regard, Khuzestan province is one of the main endemic areas for cutaneous leishmaniasis in Iran, which needs attention (3). Cutaneous leishmaniasis accounts for most cases of the disease. There are two types of cutaneous leishmaniasis in Iran, including rural cutaneous leishmaniasis caused by Leishmania major and urban cutaneous leishmaniasis caused by L. tropica (4). Several factors including the absence of a competent vaccine, vector control problems, the toxicity of antileishmanial drugs and their low effectiveness have resulted in treatment failure and worldwide leishmaniasis distribution (5-8).
As the gold standard, the Leishmania parasite is diagnosed by Giemsa staining, microscopic examination, and culture of samples collected from suspected lesions. However, such methods can only determine the parasite genus and are unable to specify different species/strains of the parasite (9-11). Differentiation between L. tropica and L. major was not accurately possible based on microscopic examination until the development of biochemical, molecular, and immunological evidence-based internal factors (12, 13). Nowadays, DNA-based methods are very popular for the detection of leishmaniasis. The main advantage of these methods is their ability to discriminate different parasite strains and detect infections even with a low number of parasites (14, 15).
Nested PCR is a useful tool in the diagnosis of leishmaniasis with high sensitivity and specificity (16, 17). However, most recently, a simple and sensitive method named loop-mediated isothermal amplification (LAMP) (18) has been widely used for the detection of bacterial, viral, protozoan, and fungal species (19, 20) and successful results have been obtained in the detection of different parasites causing infections (21, 22). The LAMP is a simple method introduced by Notomi et al. in 2000. In this method, DNA is amplified rapidly and specifically in constant-temperature condition (isothermal) (18). Moreover, it utilizes four different primers (two internal primers and two external primers) to detect six distinct regions of the target gene. Furthermore, amplification of the target region can be completed in a single step using a DNA polymerase capable of strand displacement at a temperature of 64°C (18).
2. Objectives
Given that PCR is a time-consuming and complex method, LAMP can be a simple and quick candidate to replace the Nested PCR. To the best of our knowledge, there was no study evaluating the sensitivity and specificity of LAMP for the diagnosis of cutaneous leishmaniasis in Khuzestan province, southwest of Iran. Therefore, the current survey was conducted to evaluate the utility and performance of LAMP in the diagnosis of cutaneous leishmaniasis and compare it with conventional nested PCR in the region.
3. Methods
3.1. Subjects
We enrolled 75 patients with active cutaneous lesions suspected to cutaneous leishmaniasis referring to health care centers of Khuzestan, affiliated to Ahvaz Jundishapur University of Medical Sciences to confirm their diagnoses in 2015 - 2016 (Figure 1). Clinical samples were collected from lesions of the patients according to a standard protocol. Giemsa staining was performed and the slides underwent a microscopic examination as a gold standard to be classified as 1+, 2+, 3+, and 4+, based on the number of amastigotes per microscope high-power field (23). The samples were further examined to detect parasites by both Nested PCR and LAMP methods. The demographic data were collected using a questionnaire or extracted from the patients’ medical records, as summarized in Table 1.
| Variables | No. (%) |
|---|---|
| Age group, y | |
| 0 - 10 | 30 (40) |
| 11 - 20 | 15 (20) |
| 21 - 30 | 10 (13.33) |
| 31 - 40 | 8 (10.66) |
| > 41 | 12 (14.21) |
| Gender | |
| Male | 52 (69.33) |
| Female | 23 (30.66) |
| Residence | |
| Urban | 46 (61.33) |
| Rural | 29 (38.66) |
| Number of lesions | |
| One | 45 (60) |
| Two | 23 (30.66) |
| Three | 4 (5.33) |
| ≥ Four | 3 (4) |
3.2. DNA Extraction
DNA was extracted from aspirates obtained from lesions of the patients or from the parasite culture in the case of promastigotes of L. major (MHOM/IR/75/ER), L. tropica (MHOM/IR/02/Mash10), and the virulent RH strain of Toxoplasma gondii as controls, obtained from the Tehran University of Medical Sciences, Tehran, Iran, after two days of incubation using the QIA DNeasy blood and tissue kit (Qiagen, Hilden, Germany) following the manufacturer’s instruction (4, 24). The obtained DNA was eluted in a final volume of 60 μl of AE buffer (Qiagen, Hilden Germany) and kept at -20°C until use.
3.3. Nested PCR
The first round of nested PCR was done using two primers CSB1XR (ATTTTTCG / CGA / TTTT /CGCAGAACG) and CSB2XF (C / GA / GTA / GCAGAAA C / TCCCGTTCA). The sequence of primers and reaction conditions are summarized in Tables 2 and 3, respectively. In this step, the PCR mixture contained 2 µL of the extracted DNA (100 - 150 ng/reaction), 0.4 U of Red Hot Taq (Advanced Biotechnologies, Leatherhead, United Kingdom), 75 mM Tris-HCl (pH 9.0), 2 µL of each of CSB2XF and CSB1XR primers, Tween, and 20 mM (NH4)2SO4 in a final volume of 20 µL. Amplification was done with an initial denaturation step at 94°C for 5 minutes, followed by 30 cycles of 94°C for 30 seconds, 55°C for 60 seconds, and 72°C for 90 sECONDS using an Eppendorf thermocycler (Eppendorf AG22331, Hamburg, Germany). Then, 2 µL of the diluted product (one-tenth dilution in distilled water) obtained from the first-round was used as a template for the second round in an overall volume of 20 mL under the same conditions as those for the first round, except for using primers 13Z and LiR. Eventually, the PCR products of the second-round were electrophoresed on a 2% agarose gel.
| Name of Primer | Sequence (5 - 3) |
|---|---|
| CSB2XF | CGA GTA GCAGAAA C TCCCGTTCA |
| CSB1XR | ATTTTTCG CGA TTTT CGCAGAACG |
| 13Z | ACTGGGGGTTGGTGTAAAATAG |
| LiR | TCGCAGAACGCCCCT |
| Step | Cycle | Temperature, °C | Time(s) |
|---|---|---|---|
| Initial Denaturation | 1 | 95 | 300 |
| Denaturation | 24 | 94 | 60 |
| Annealing | 58 | 120 | |
| Extension | 72 | 120 | |
| Final Extension | 1 | 72 | 300 |
3.4. Loop-Mediated Isothermal Amplification (LAMP)
The LAMP method was adopted from the literature (25, 26) with subtle modifications. The reactions were performed using a set of primers targeting 18S rRNA coding the sequences of Leishmania parasites. The sequence of primers is summarized in Table 4. Each reaction mixture was prepared in a total volume of 25 µL containing 1.4 mM of each deoxynucleotide triphosphate, 5 pmol each of F3 and B3c primers, 40 pmol each of FIP and BIP primers, 20 mM Tris-HCl, 0.8 M betaine, 10 mM (NH4)2SO4 (pH 8.8), 10 mM KCl, 0.1% TritonX-100, 8 units of Bst DNA polymerase large fragment (New England Biolabs, Ipswich, MA), 8 mM MgSO4, 0.004% malachite green (MG) dye (diluted in distilled water), and 2 µL of sample DNA. The resulting mixture was incubated at 64°C for 60 minutes with heat block. Then, the mixture was chilled on ice to visualize the precipitation. Moreover, for further validation, 7 µL of the LAMP products were electrophoresed with a 100-base pair DNA size marker (New England Biolabs) on a 2% agarose gel and visualized under ultraviolet light after staining with a DNA safe stain dye. Furthermore, instead of the extracted DNA, the control tube contained distilled water and the DNA of the standard species of L. major (MHOM/IR/75/ER), L. tropica (MHOM/IR/02/Mash10), and the virulent RH strain of T. gondii.
| Name of Primer | Sequence (5’ - 3’) |
|---|---|
| F3-Le.rRNA | GGGTGTTCTCCACTCCAGA |
| B3-Le.rRNA | CCATGGCAGTCCACTACAC |
| FIP-Le.rRNA | TACTGCCAGTGAAGGCATTGGTGGCAACCATCGTCGTGAG |
| BIP-Le.rRNA | TGCGAAAGCCGGCTTGTTCCCATCACCAGCTGATAGGGC |
3.5. Statistical Analysis
One-tailed t-test was used to indicate differences in sensitivity and specificity at a significance level of P < 0.05. The true-positive (TP), true-negative (TN), false-positive (FP), and false-negative (FN) results were determined. The sensitivity, specificity, and accuracy of the methods were also calculated. Statistical analysis was done using SPSS 16.0 (SPSS Inc., Chicago, IL, USA).
4. Results
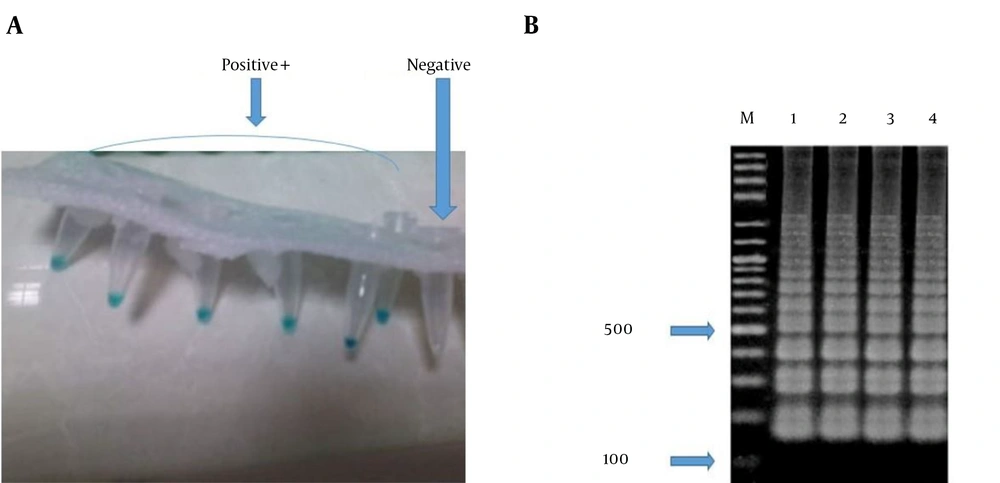
All samples that were microscopically positive (75) were also positive by both Nested PCR and LAMP methods. The sensitivity of LAMP for cutaneous leishmaniasis samples was 98% and its specificity was 100%. The sensitivity and specificity of kinetoplast minicircle DNA (kDNA) Nested-PCR were 100%. In both Nested-PCR and LAMP, kDNA and 18S rRNA coding sequence of Leishmania parasite were successfully amplified, respectively, by the primers in sample tubes containing each target DNA (Figures 2 and 3). The results obtained from both thermal-cycler and heating block were similar. Moreover, we investigated the sensitivity of the LAMP assay using serial dilutions of L. major DNA (104 to 0.1 parasites). The best detection occurred in the highest number of parasites (104) although the LAMP assay could also recognize parasites in low numbers (0.1 parasites per µL). According to our findings, no proliferation was observed in negative control samples.
Colorimetric LAMP sensitivity and agarose gel electrophoresis detection. A, Malachite green (MG)-based LAMP sensitivity reaction products observed by the naked eye without a UV illuminator after incubation. Light blue: positive reaction; colorless: negative reaction. B, Detection of L. major DNA by LAMP using electrophoresis on a 2% agarose gel, followed by DNA safe staining: lane 1,: positive control of L. major (L. major MHOM/IR/75/ER); lanes 2 - 4, typical ladder-shaped pattern of a positive reaction. M, 100-bp DNA size marker.
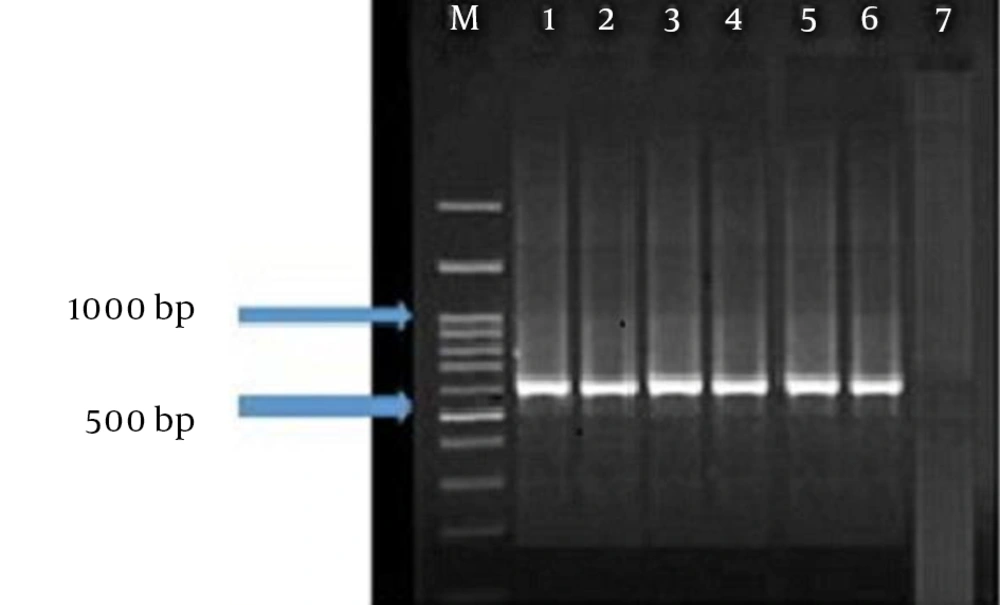
The results of LAMP assay were easily detectable by visual inspection. All specimens containing DNA of the Leishmania parasite appeared in blue while T. gondii samples and samples obtained from healthy humans were colorless (Figure 2A). The results of gel electrophoresis also confirmed successful amplification by the LAMP assay (Figure 2B). Furthermore, all the samples were tested by the Nested PCR technique. The results obtained by gel electrophoresis of the Nested PCR products compared to the electrophoresis banding patterns of standard samples showed that all of the 75 isolates were identified with a 560-bp length fragment attributed to the L. major parasite (Figure 3).
5. Discussion
The number of patients with cutaneous leishmaniasis is increasing worldwide, especially in developing countries. A cost- and time-effective diagnosis could help better control and efficiently treat leishmaniasis. However, current diagnostic methods are time-consuming and require advanced equipment. Thus, the simplification of detection methods is critical in this field. Since leishmaniasis is an important health-related issue in Khuzestan province, southwest of Iran, the present study aimed to evaluate the usefulness and performance of LAMP in the diagnosis of cutaneous leishmaniasis compared to conventional Nested PCR in the region. The microscopy is the gold standard for detection of parasites. Although microscopy is highly sensitive, it has low specificity. Recently, the PCR method is of interest because of high sensitivity and specificity (18, 19, 22).
Khuzestan province is one of the endemic areas of cutaneous leishmaniasis in Iran, with the dominance of L. major (3). In this study, all samples were diagnosed as positive by the direct microscopic examination and the parasite species were determined to be L. major by Nested PCR. In previous studies, similar methods were used to diagnose leishmaniasis and showed that L. major is responsible for cutaneous leishmaniasis in this region (4, 10, 22, 23, 27). As previously mentioned, in this study, we aimed to compare the specificity and sensitivity of Nested PCR and LAMP tests for cutaneous leishmaniasis detection. In other words, we intended to investigate how many positive slides by direct microscopic examination were positive separately by Nested PCR and LAMP. Accordingly, the Nested PCR was conducted using the primers targeting kinetoplast minicircle DNA of Leishmania parasites as described in the literature. However, in the case of LAMP, because the conserved regions are needed and the minicircles are highly variable, we performed the LAMP assay using primers targeting the 18S ribosomal RNA gene according to Sriworarat et al. (28-31).
Our study showed that LAMP can be used widely for detecting L. major because it is a simple and low-cost technique that does not need complex tools and, most importantly, the final result of this method can be visualized easily. Furthermore, the LAMP acts highly specifically because of using specific primers, which makes it again superior to other techniques (18). Some previous studies investigated the specificity and sensitivity of LAMP compared to other methods. Takagi et al. (32) reported the specificity and sensitivity of LAMP compared to Nested PCR in the diagnosis of L. donovani and showed that the sensitivity of LAMP was 10 times the sensitivity of Nested PCR while both methods were specific. In line with this study, our result also confirmed LAMP as a good alternative for Nested PCR (32).
Similar to our work, Kothalawala and Karunaweera (33) recently investigated the potential utility of LAMP to diagnose cutaneous leishmaniasis in Sri Lanka. They enrolled 31 cutaneous leishmaniasis patients who had been diagnosed clinically. They found that LAMP was positive for 82.6% of microscopically positive samples. Moreover, their results showed that the specificity of the LAMP was 100% while the time and cost reduced by 50% when the LAMP was used instead of Nested PCR. These findings confirm our results (33). Nzelu et al. (25) in Peru established a study to determine the diagnostic value of LAMP for rapid diagnosis of Leishmania DNA from tissue samples spotted on an FTA card. They enrolled 120 patients suspected to cutaneous leishmaniasis of whom, 71 patients were detected with cutaneous leishmaniasis. Contrary to our results, they found that LAMP had a high diagnostic sensitivity down to near 0.01 parasites per µL. Moreover, their findings showed that the FTA-LAMP assay was as proficient as the nested PCR method using an FTA card as a template. In accordance with our work, they showed that pre-added malachite green enabled the visual discrimination of results by the naked eye (25).
Adams et al. (34) conducted a study to evaluate LAMP for cutaneous leishmaniasis and visceral leishmaniasis diagnosis. To this end, lesion swab samples were obtained from 105 patients suspected to cutaneous leishmaniasis and blood samples were collected from 50 suspected visceral leishmaniasis patients. The diagnostic accuracy was calculated compared to the results of the microscopic examination as a gold standard. Moreover, the specificity of LAMP was obtained using samples of other pathogens including malaria parasites, arboviruses, and bacteria. They found that the LAMP assay had 95% sensitivity and 86% specificity for the diagnosis of cutaneous leishmaniasis. In the case of visceral leishmaniasis suspected samples, the sensitivity and specificity of the LAMP assay were 92% and 100%, respectively. These results again confirm our findings and proposed the LAMP assay as a quick, one-step, single-tube, and highly sensitive method for detection of cutaneous leishmaniasis (34).
Ghasemian et al. (26) conducted a study to compare Nested PCR and LAMP for diagnosis of visceral leishmaniasis in 47 samples. Consistent with our findings, they showed similar sensitivity and specificity for both tests; however, considering that LAMP was simpler, easier, time-effective, and cost-effective (26), it was proposed as a suitable alternative for Nested PCR. The same result was obtained by Verma et al. in India (New Delhi) (35). They developed a LAMP assay based on L. donovani kDNA for detection of L. donovani, L. tropica, and L. major infections in biopsy samples collected from people with cutaneous leishmaniasis, visceral leishmaniasis, and post-kala-azar dermal leishmaniasis (PKDL) using a closed tube to prevent cross-contamination.
The test identified the parasite in 64 of 66 (96.97%) visceral leishmaniasis blood samples, 15 of 15 (100%) visceral leishmaniasis bone marrow aspirate samples, and 65 of 67 (97%) PKDL tissue biopsy samples. In addition, the assay was assessed for cutaneous leishmaniasis diagnosis where it was established positive in 8 of 10 (80%) tissue biopsies (35). Another study was conducted by Fallahi et al. to compare Nested PCR and LAMP for diagnosis of T. gondii DNA in blood samples of children with leukemia. To this end, they used primers targeting the RE (repeated element) and B1 genes. Their results showed that the LAMP had higher sensitivity and accuracy than Nested PCR. Accordingly, they found that 92% and 86% of the samples were positive using LAMP targeting the RE and B1 genes, respectively, while using Nested PCR targeting the RE and B1 genes, only 82% and 68% of the samples were positive, respectively (36). Using T. gondii DNA in our study, we confirmed the specificity of LAMP for diagnosis of L. major. However, a limitation to previous studies was the lack of using the DNA of T. gondii for evaluation of cross-reactivity.
5.1. Conclusions
The sensitivity of the LAMP method is similar or even higher than the sensitivity of Nested PCR. Given the convenience, low-cost, and no need for complex tools, the LAMP method can be used as an available alternative for Nested PCR in the diagnosis of cutaneous leishmaniasis.