1. Background
There is a wide range of microbial inhibitors or lethal agents with a peptide-based identity, which are known as antibacterial peptides. These peptides are at the forefront of controlling microbial agents and are considered as an integral part of the immune system of eukaryotes and prokaryotes. Antimicrobial peptide (AMP) production occurs in the face of any infections and is secreted by stimulation and induction, depending on the pathogen involved. They typically have the characteristics including small in length (12 to 50 amino acids), amphipathicity, and positively charged subunits. The presence of such properties allows them to penetrate the cell wall or negatively charged cell membranes of pathogens (1-3).
Anti-microbial peptides act either directly upon the degradation of the bacterium’s cell membrane or by regulating and stimulating other elements from the cell environment or innate immune system. The increased resistance to antibiotics is permanently a considerable concern (4, 5). Therefore, natural antibiotics or those derived from nature have become more popular since they provide the highest harmony with the body and give the chance to withstand the least possible disturbance factors. Meanwhile, the identification and use of antimicrobial peptides today are more considered compared to other effective treatment candidates (6). Depending on the type of subunit and molecular structure, AMPs are divided into four categories including (1) anionic peptides that are richly found in the subunits of glutamine and aspartic acid; (2) linear cationic peptides with alpha-helix structure and without cysteine in their amino acid sequence; (3) cationic peptide fulfilled with specific amino acids, including proline, arginine, phenylalanine, glycine, and tryptophan; and (4) anionic and cationic peptides with cysteine in their sequences and containing one up to three disulfide bonds in their structure and bactenecin is classified in this category.
Heretofore, more than 2,420 antimicrobial peptides have been identified, of which 1,819 ones are isolated from animals. Although these peptides are often expressed as pro-peptides, some of them have a permanent expression and some are expressed by induction, e.g., by inflammation or ulceration (5, 6). Bovine dodecapeptide or bactenecin is an antimicrobial peptide isolated from cow’s neutrophil cells (7). Bactenecin is one of the smallest cationic antimicrobial peptides, which is identified with 12 amino acids. Regardless of the environment in which the peptide is located, bactenecin is recognized with a beta-turn or loop-like structure due to the presence of a disulfide bridge from two cysteines in its sequence at positions 3 and 11. In the case of bactenecin, mild activity against Gram-negative bacteria has been identified and the presence of cysteine in its sequence over the antimicrobial activity of bactenecin has also been shown (7, 8). Additionally, earlier studies have not implied the stronger antibacterial effects of modified peptides against Gram-negative bacteria (9, 10).
Besides, as mentioned before, the most known factors involved in the function and activity of cationic antimicrobial peptides include positive charges, hydrophobicity, and amphipathicity. Also, in AMPs like bactenecin, the conformation of disulfide bridges or loop structures is essential for stabilizing alpha/beta-sheet structures (11, 12). Hence, the present study aimed at investigating the bactenecin bactericidal activity and determining the effect of increasing the positive charges of amine and carboxyl terminus and increasing the hydrophobicity of the central part of the antimicrobial peptide.
2. Objectives
We studied the effect of both factors on peptide anti-bacterial activity. Also, to preserve the native loop structure of bactenecin, neither removal nor increase in the number of cysteine residues was performed. Through this investigation, we intended to determine the effectiveness of these changes on bactenecin, especially against Escherichia coli.
3. Methods
3.1. Reagents and Strains
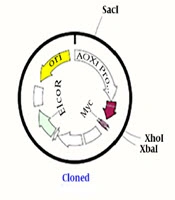
Escherichia coli DH5α, Pichia pastoris GS115, and Escherichia coli ATCC 25922 were the strains used for plasmid amplification, gene manipulation, and protein expression, respectively, to conduct antibacterial activity assays of peptides obtained at our laboratory (NIGEB, Iran). In addition, the pPICzαB plasmid was used for the expression of exogenous peptides preserved at our laboratory. Restriction endonucleases XbaI, XhoI, EcoRI, SacI, and other enzymes were used for all processes from cloning to expression obtained from Fermentas Thermo Scientific (Glen Burnie, MD, USA).
The kits for DNA purification and plasmid extraction were purchased from Roche (Basel, Switzerland). All chemical reagents and culture media with high purity were prepared from Sigma Aldrich (St. Louis, Missouri; United States). DEAE and SP-sepharose were packed as columns at the time of use; they were purchased from GE Healthcare (Piscataway, NJ).
3.2. In Silico Study
3.2.1. Design and Synthesis of the Gene Encoding Native and Mutated Peptides
About 20 designed derivative candidates of bactenecin (BN) were checked using the APD3 database to calculate their functionality extent as anti-microbial peptides (13). We selected three candidates characterized by the increased C-terminal positive charge (BM1), higher hydrophobicity in the loop part of the peptide (BM2), and a more elevated amount of positive charge and hydrophobicity in both regions (BM3), all of which were predicted by advanced antimicrobial possibility. Subsequently, their physicochemical properties were rechecked with the ExPASy PI/MW tool (14).
For structural studies and initial simulation of peptides, the ab initio simulation method was used through the PEP-FOLD server (15). Since the molecular dynamics technique is a remarkable method for studying the structure and intercellular construction of molecules and because there was initially no 3D model for bactenecin, the peptide structural sequence was drawn with ChemSketch Ver. 12.01 software; then, Open Babel GUI Ver. 2.3.2 software was used to generate PDB formatted files (16) and finally, the data were prepared for molecular dynamics studies.
Molecular dynamics simulation of peptides was performed in water by the OPLSA force field from GROMACS Ver. 4.6.5 computational package series. Peptides’ positive charges were neutralized by adding chlorine ions to the simulation system. In this case, due to the size difference between peptides, the comparison of the obtained data was made only between BM1 and BM2 to analyze the effect of the elevation of positive charges and hydrophobicity.
3.2.2. Cloning, Transfection, and Expression in Pichia pastoris GS115
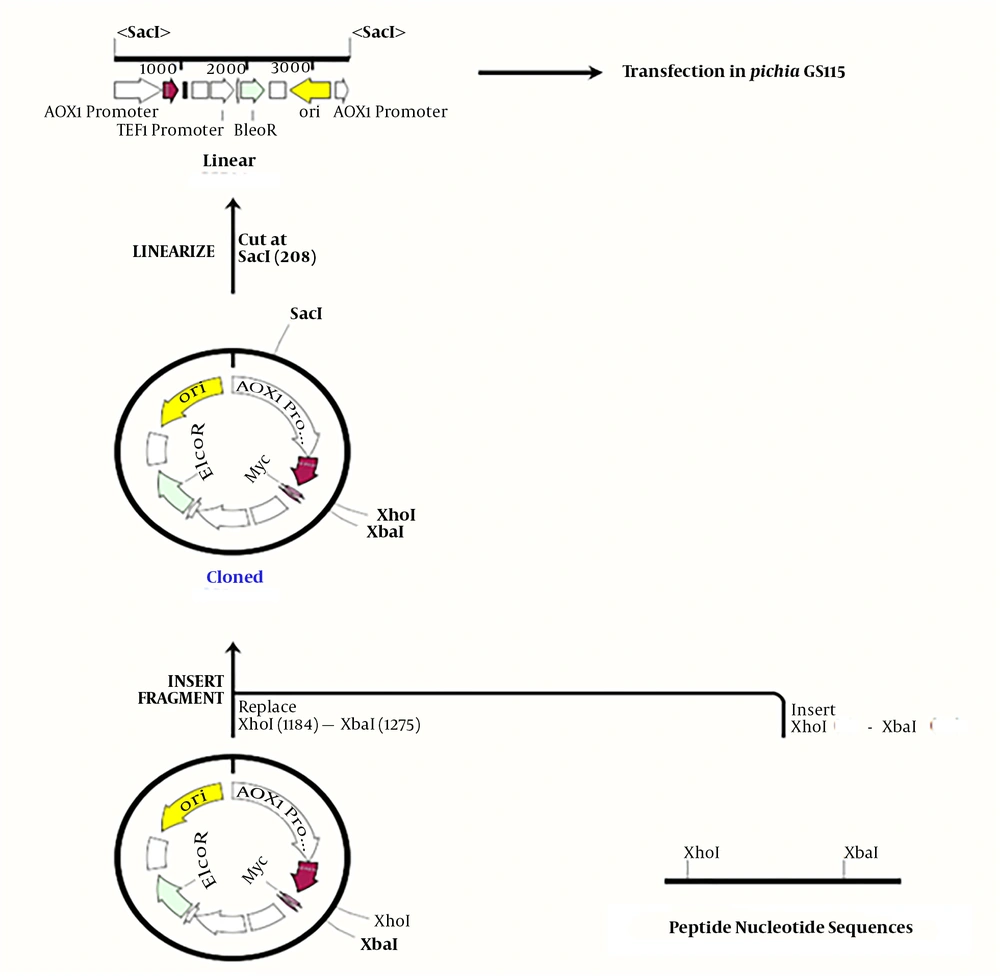
In this study, Pichia pastoris GS115 was used as a host for obtaining better expression. Each peptide nucleic acid sequence was optimized based on codon usage in this yeast (17, 18). In order to facilitate the cloning of peptides’ nucleic acid sequences (due to the short sequence size of each peptide and difficult detection of cloned ones on the agarose gel in the expression vector), a tail protein sequence with an EcoRI restriction recognition site at both ends of this protein was attached as a helper sequence. Restriction enzyme recognition sites at each side of peptide sequences were designed and introduced, as follows: XhoI restriction recognition site to upstream, two stop codons at the end of the sequence, and XbaI restriction recognition site at the downstream of every peptide nucleic acid sequence (19).
Previous studies have shown that native bactenecin has no effect on yeast cells, but as peptide derivatives were designed and expressed for the first time, so their action on yeast cells were ambitious for us either. In this study their effect on yeast cells were analyzed and no effect were mentioned. The following items were introduced into the vector: each fragment containing the desired peptide from BN, BM1, BM2, and BM3 with the helper sequence, the pPICzαB expression vector digested with XhoI and EcoRI, and the entire fragment. After reassuring the cloning, the manipulated vectors from pPICzα-BN to pPICzα-BM3, which were digested by the XbaI restriction enzyme, as well as the additional parts, were removed; finally, only remained the desired peptide sequence in the vector. Escherichia coli DH5α was used as a cloning host for the elevation of recombinant plasmids copy numbers. Cloning accuracy was checked by AOX universal primers and the cloned yeast genome extracted from each sample was sent for sequencing.
To introduce an expression vector containing the nucleic acid sequence of desired peptides to competent Pichia GS115 cells after the linearization of recombinant and control vectors with SacI, the Bio-Rad gene-Pulser electroporation system was used at a voltage of 1500 V, a capacitance of 25 µF, and a resistance of 200 Ω. After two hours of preservation in a 1 sorbitol solution at a temperature of 28ºC, transfected yeasts were cultured in the YND plates containing zeocine as a selective antibiotic. After 48 hours, the colonies were re-cultured in MMH and MDH plates to isolate Mut+ and Muts phenotypes following the Invitrogen pPICZα instructions (tools.thermofisher.com/content/sfs/manuals/ppiczalpha-man.pd).
Mut+ colonies were re-cultured and their genomes were extracted by the LioAc-SDS method to verify the accuracy of cloning. All derived genomes were checked by universal AOX primers and then were sequenced to isolate positive clones (20, 21). For further expression of target peptides, Pichia GS115 cells were induced by methanol to a final concentration of 0.5% for 72 hours at a temperature of 28ºC. Then, 2% histidine and biotin were added as supplementary to each one liter of YNB medium. Due to the presence of alpha protein secretion signal at the upstream of each nucleic acid sequence of peptides (checked in every step of cloning), after 48 hours of cultivation at a temperature of 30ºC, the supernatant was isolated from cultured yeasts in three liters of the medium at a speed of 12000 rpm and then filtered through a cellulose acetate membrane.
3.2.3. Sample Concentration and Partial Purification of AMPs by Ion Exchange Chromatography
After the calculation of PI for each peptide, ion-exchange chromatography was performed on the filtrated supernatant of every sample to raise the peptides’ concentration. With this method, not only was the extra color removed from samples, but also peptides or other proteins were eliminated from the opposite charge within the test. The net charge on a protein is zero at the isoelectric point (pI), positive at pHs below the pI, and negative at pHs above the pI. Therefore, the specimens reached a pH of 7.5; then, they were incubated at a temperature of 4ºC for 30 min and centrifuged at a speed of 12000 rpm for 5 min. The supernatants were collected and then passed through DEAE, an anion exchange column; also, unbound fractions were completely collected. Next, the pH of equilibrated fractions raised to 12.5 and crossed over the SP-sepharose cation exchange column. The column was washed by a buffer containing 50 mM Tris and 250 mM NaCl. The outlet was collected by Bio-Rad fraction collector and their absorbance was scanned in a range of 200 - 300 nm. Fractions with absorbance in these wavelengths were collected, filtrated, and lyophilized for the next step.
3.2.4. HPLC Analysis of Partially Purified AMPs
The partially purified antimicrobial peptides were further purified by a reparative reversed-HPLC (RP-HPLC) on a C18 column (250 cm, 4.6 mm, 5 µm) (Azura, Knauer, Germany). This column was pre-equilibrated using the mobile phase of acetonitrile (80% v/v with 20% water). Attached peptides were eluted with a gradient of acetonitrile and a flow rate of 1.0 mL per min; also, for better resolution, injection volumes were reduced to 200 µL. Eluted absorbance was monitored at wavelengths of 210, 220, and 250 nm.
3.2.5. LC-Mass Analysis of AMPs
For purity determination and identification of peptides, the LC-mass equipment (Agilent, 6410QQQ, CA, USA) was used with a measurement accuracy of ng/µL. From each sample, a fraction was gathered that acted more potently against bacteria compared to the positive control. The positive fractions containing our required peptides were identified and the purity rate of each peptide was determined by HPLC at a wavelength of 214 nm.
3.2.6. Minimum Inhibitory Concentration (MIC) Assay of AMPs
Minimum inhibitory concentration tests were conducted for 48 hours using the broth dilution microtiter method with serially diluted concentrations of BN, BM1, BM2, and BM3 in 96-well plates. Escherichia coli (ATCC 25922) was used as the bacterial strain in this study. It was employed as a representative of Gram-negative cells. To evaluate the antimicrobial activity of bactenecin and derivative candidates, the bacterial strain was cultured in Luria broth (containing 10 g bacto-peptone, 10 g NaCl, and 5 g yeast extract per liter) with the optical density (OD) of 600 nm reaching 0.08 - 0.1 as 0.5 McFarland. As a reference medium for MIC analysis, Moller Hinton medium was used according to the NCCLS standards (22-24).
3.3. Statistical Analysis
For MIC data analysis and comparison between controls and peptide samples, IBM SPSS Ver. 24 Software was used. The data were interpreted based on the average of at least three repetitions for each sample and they were expressed as mean ± standard deviation. The statistical significance of the difference between samples and controls was checked out by the analysis of variance (ANOVA), followed by the Duncan test. For significance evaluation, the P value was considered to be less than 0.01 (P < 0.01).
4. Results
4.1. Structure Prediction and Cloning
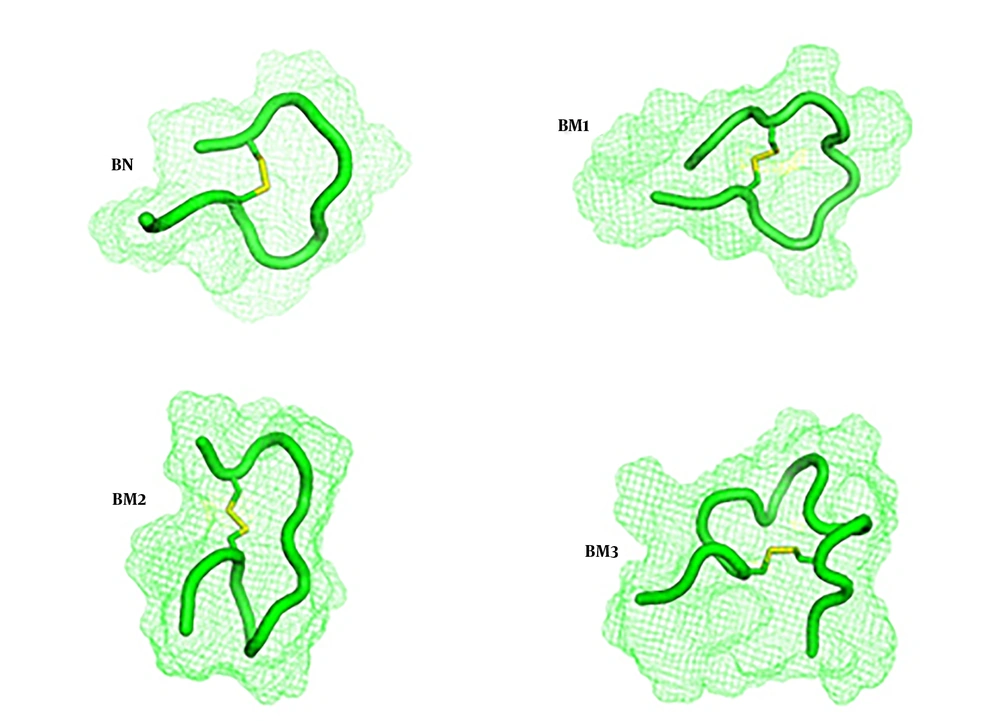
As the key amino acid cysteine was not changed, the anticipated structures from the molecular dynamics study were the same as what predicted through the PEP-FOLD server and they had loop structures, as expected (Figure 1). Disulfide bridges were formed during the molecular dynamics study and were shown with yellow bars. As conventional for all four peptides, the designed cloning platform was checked by SnapGene Ver. 1.1.3. Software (Figure 2). Sequencing results confirmed the accuracy of cloning and correct placement of peptides’ nucleic acid sequences after pPICzαB secretion signal. The calculated physicochemical features of peptides were collected and presented in Table 1 and ion-exchange chromatography was used for their semi-purification.
| Peptide | Peptide Sequences | Amino Acids Qty | PI | MW | Charge | Hydrophobicity Rate, % |
|---|---|---|---|---|---|---|
| BN | RLCRIVVIRVCR | 12 | 11.53 | 1485.92 | +4 | 66 |
| BM1 | RLCRIVVIRVCRRR | 14 | 12.00 | 1798.29 | +6 | 57 |
| BM2 | RLCRIVIVVIRVCR | 14 | 11.53 | 1698.21 | +4 | 71 |
| BM3 | RRLCKRIVVVIRKVCRRR | 18 | 12.13 | 2309.96 | +9 | 50 |
4.2. Determination and Characterization of Peptides
Whole observed peaks for BN, BM1, BM2, and BM3 compared to the control sample were fractionated over time. Therefore, as the primary step to separate positive fractions after HPLC, we examined them with the same conditions in the 96-well plate only for the detection of bactericidal or bacteriostatic properties (Table 1).
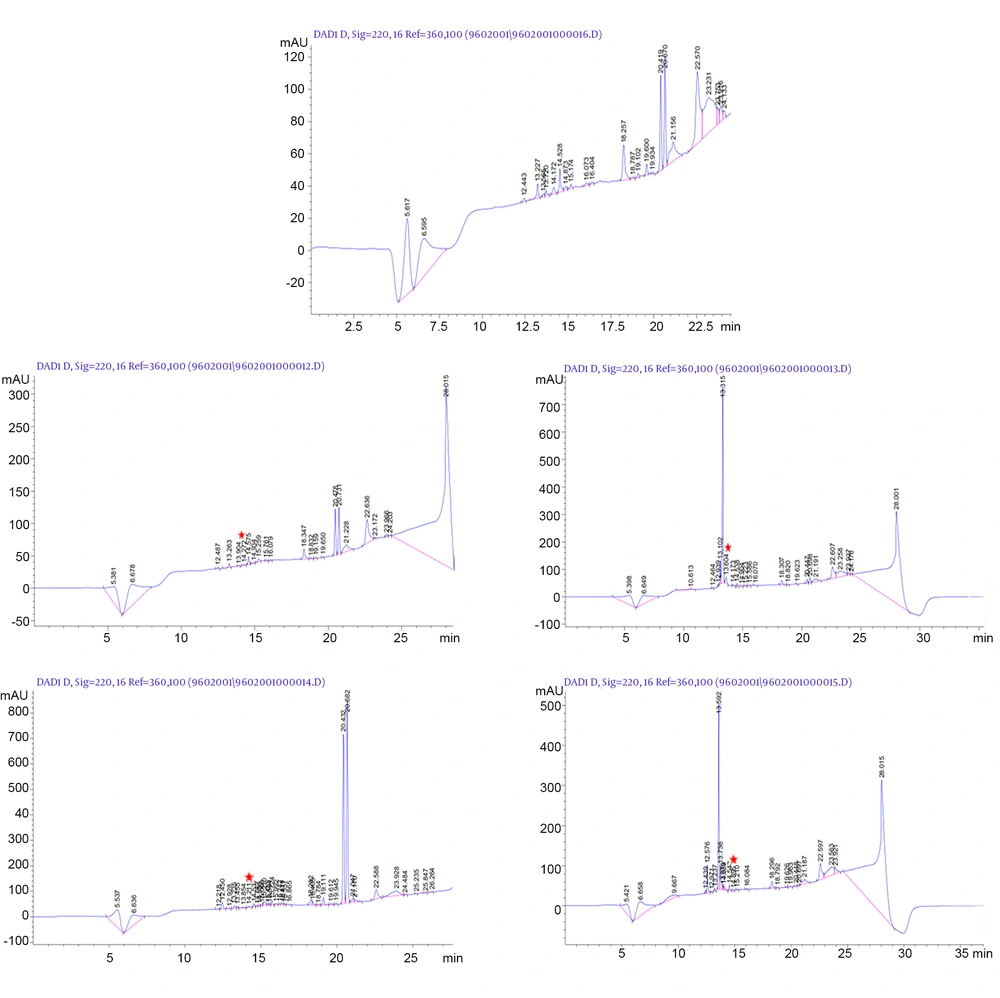
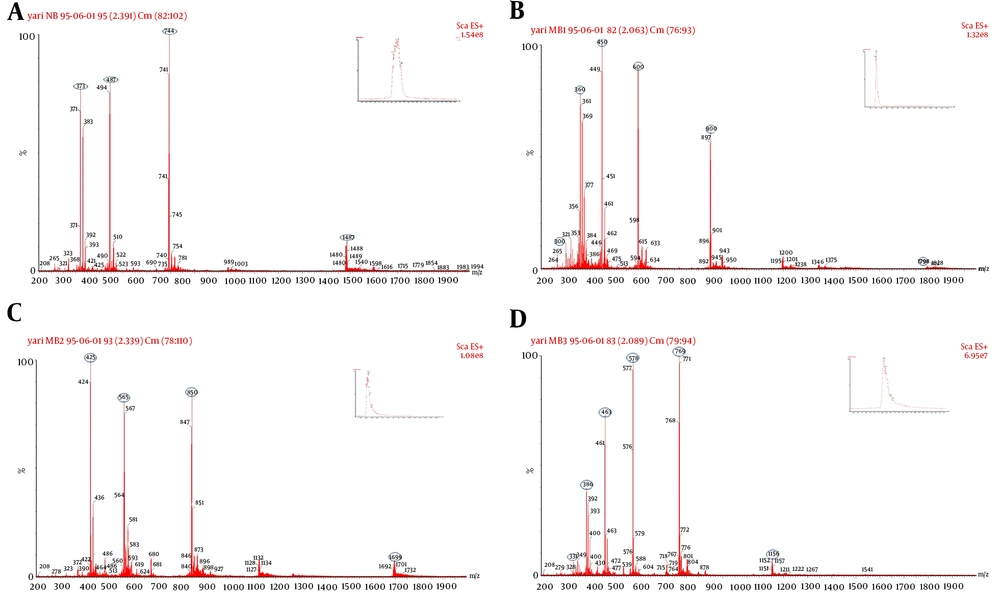
The fractions with the ability to make delay or inhibition in E. coli growth were isolated, lyophilized, and sent to LC-mass for identification. Figure 3 shows the peaks observed in HPLC from control to BM3. In contrast to the control, the fractions not observed in peptide supernatants were collected. The probability of every fraction functionality was examined. For each peptide, the time during which functional fraction was isolated is shown with the red star in Figure 3. The retention times of each peptide to pass through the HPLC column were 14.27, 13.60, 14.21, and 14.89 min for BN to BM3, respectively. The samples were sent for mass spectrophotometry and thus all peptides were characterized (Figure 4). The purity rate was determined to be 70%, according to their absorbance at a wavelength of 214 nm.
Lc-mass analysis result of positive fractions which collected from HPLC. Mass diagram of each peptide indicated their capacity of ionization through mass spectrophotometry. BN with the positive charge of 4 has 4 capacities of ionization that indicated in A, as follows BM1 with 6; B, BM2 with 4 positive charges; C, were completely detected. Because of the molecular weight higher than 2000 dalton which was not detectable with our LC-mass apparatus, BM3’s some ionization masses not detected but other ones were shown; D, oval shapes specify the ionized masses.
Their dry weight was used to determine peptides’ yields. BM2 had the lowest expression with a yield of 190 µg/L in the cultured yeast, BM1 and BN had a relatively similar yield of 210 µg/L, and better expression was achieved for BM3 with a yield of 240 µg/L. Another remarkable result obtained from mass data was that no amino acid was added to the peptide sequences (which is essential for their function), indicating the full expression and secretion in the yeast system in the right way.
4.3. Minimum Inhibitory Concentration of AMPs
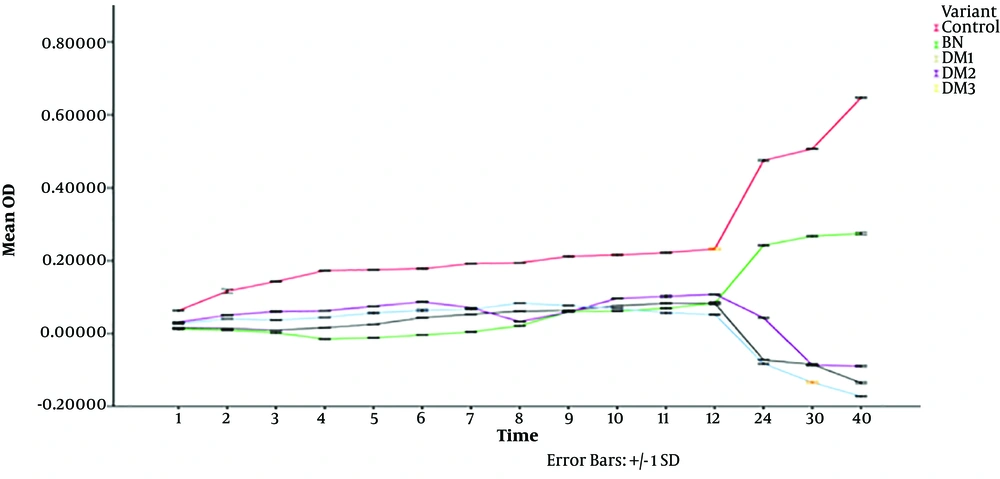
After defining the identity of the peptides, the MIC standard test was conducted within the dilution range of 0.5 - 32 µg/mL in 96-well plates and the bacterial growth was tracked over time by an ELISA reader at a wavelength of 600 nm. The functional MICs of BN, BM1, BM2, and BM3 peptides against E. coli were determined. The results are presented in Table 2, which implied that the modified peptides better acted against E. coli. Statistical analysis of peptides compared to the control showed significant differences; Statistical analysis of peptides to control showed significant values; as a result, the impact of sequence altering in entirely three forms from only elevated positive charge and high hydrophobicity to elevation in both features through time was more consistent in contrast with native bactenecin. The statistical results are presented in Table 3 with significant values. SPSS results are shown in a graph in Figure 5.
| Organism’s Peptides | MIC, µg/mL | |
|---|---|---|
| 50% | 90% | |
| Escherichia coli (ATCC 25922) | ||
| BN | 8 | ND |
| BM1 | 2 | 4 - 8 |
| BM2 | 4 | 32 |
| BM3 | 2 | 8 |
| Variant (I) | Variant (J) | Mean Difference (I-J) | Std. Error | Sig. | 99% Confidence Interval | |
|---|---|---|---|---|---|---|
| Lower Bound | Upper Bound | |||||
| BN | Control | -0.17791556c | 0.02079784 | 0.000 | -0.2409463 | -0.1148848 |
| BM1 | Control | -0.23280000c | 0.02079784 | 0.000 | -0.2958307 | -0.1697693 |
| BM2 | Control | -0.20262222c | 0.02079784 | 0.000 | -0.2656530 | -0.1395915 |
| BM3 | Control | -0.23067556c | 0.02079784 | 0.000 | -0.2937063 | -0.1676448 |
aDunnett t-tests treat one group as a control and compare all other groups with it.
bThe dependent factor in this test was optical density.
cThe mean difference is significant at the 0.01 level.
5. Discussion
The native form of bactenecin showed a rapid and abrupt response that underpinned its role in neutrophils as the first actor in the innate immune system. This study simultaneously depicted well the bactenecin performance in the innate immune system, which may be related to the rapid effect of this peptide, even at low concentrations compared to what indicated in the MIC standard test, although a higher concentration of this peptide is required for more than 12 hours (17). For tracking the functionality of peptides in the fixed period on E. coli, 2 µg /mL of each AMP was used. Meticulously, the test was performed like MIC, where only one concentration was used. The results implicated that the natural sequence of bactenecin reacted rapidly to Gram-negative bacteria (E. coli) although the length and range of effect remained limited, which is intrinsically compatible with the function of the innate immune system. However, it should be taken into account that the act of this peptide was faster than that of its modified ones since the bacterial growth after 8 - 10 hours was evaluated to be reasonable. Besides, the incremental effect became more and more influential in the case of peptides derived from bactenecin.
Previous studies on synthetic forms of bactenecin reported the increased efficiency of this peptide with changes in its sequence and structure (25, 26). The MIC test with E. coli (ATCC 25922) was conducted here for each peptide (14-19). The MIC test result for BN proved the proper implementation and outcome of our experiments, which were similar to previous studies (about 8 µg/mL) and there seemed to be no difference between the expressing one and synthetic forms. The functional progression of BM1 and BM3 peptide variants was another important factor that revealed a comparatively similar effect on E. coli, by increasing the positive charge and hydrophobic load simultaneously. The MIC50 findings in a duration of 16 - 24 hours for BM1, BM2, and BM3 were 2, about 4, and 2 µg/mL, respectively, and were completely different from those of the native peptide.
The results of this study also pointed to the BM2 peptide function, which was accompanied only by a higher level of hydrophobicity. The poor and delayed action of BM2 compared to the other two varieties was a piece of evidence that a more considerable amount of this peptide needed for bactericidal effect. The results of peptides action against E. coli through the same concentration implied a new functional pathway for each peptide within one-hour interval monitoring even for 48 hours.
The MIC data analysis of bactenecin derivative proved that the improvement in the functionality of these peptides was more relevant to increased positive charge than hydrophobicity. Besides, a combination of charge and hydrophobicity percentage elevation was not much better than the positive charge alone. The other conclusion from these data may implicate the importance of stability of the maternal structure and cysteine residues, which need more analysis. Besides, studying the size and conformation of the loop as done by Kiricsi et al. (27) will be very useful and give us a more in-depth look at how these peptides work.
In this study, the control sample was used throughout the designed process. In a uniform method, at all stages from cloning and expression to final analysis with samples containing antibacterial peptides, the control sample was assayed, as well. At all stages, the extracted peptides from samples and empty vector-transfected yeast (control) were analyzed. After ion-exchange chromatography, semi-purification, and spectrophotometric analysis, the bacterial growth test was performed on the samples regardless of the presence or absence of peptides to remove false-positive data, which showed no antibacterial activity even in purified contents. In the later stages of HPLC, the collected fractions were again lyophilized and examined for their effects on bacterial growth. In addition, for having access to the native synthetic sample obtained from our laboratory, a comparison was made between the cloned sample containing 70% purity and the synthetic one, which showed no effect on bacterial growth in the presence of impurities.
Since it was not crucial to make any changes in the structure and sequence of the peptides, the tag was not added to the end of each peptide. The 100-nucleotide designed helper sequence was used only to aid the cloning of small peptides in the expression vector. This sequence was removed by enzymatic digestion and through ligation to completely integrate each peptide nucleotide fragment into the expression vector. Contrary to the expectation that a higher amount of peptide could be due to their better performance against bacteria, other results from this study rejected such an idea (28). For further study and improvement of the yield, peptides expression can be a solution in other hosts or considered as an inclusion body (23, 29, 30). Attributable to the high cost of peptide synthesis, the ability to express peptides under suitable conditions can be an excellent way to reduce the cost of producing peptides with medicinal properties (31). A closer look at the structure of varieties designed in this experiment and how they interact with the bacteria and other microorganisms’ cell walls are proposed for further studies.
5.1. Conclusions
The results of this study support our hypothesis that the design of peptides can increase positive charges and hydrophobicity, either alone or together, without altering cysteine amino acids based on the structure of their maternal peptide (bactenecin), aiming to enhance the activity of bactenecin against Gram-negative bacteria. BM1 that had the increased positive charge and BM3 that had simultaneously the increased charge and hydrophobicity showed stronger and faster effects than BM2, which held the mediocre result, even though BM2 with the modification only in hydrophobicity percentage registered the lowest in contrast to other modified AMPs. However, our limitations in pure peptides isolation, laboratory safety terms, and the lack of class II laminar hood did not allow us to examine pathogenic strains of Gram-negative bacteria, which suggest further experiments with more strains and Gram-positive strains.