1. Background
Worldwide, viral diarrhea is the main cause of gastroenteritis in children, being the severe acute gastroenteritis (AGE) one of the most important causes of morbidity and mortality in children under five-years-old. In 2016, despite the implementation of new prevention strategies, such as vaccination, diarrhea mortality in infants reached 446,000 cases (1). Viruses from four families Reoviridae (group A rotavirus), Caliciviridae (norovirus), Adenoviridae (adenovirus, mainly serotype 40/41, coded as species F), and Astroviridae (astrovirus) are considered major causal agents of acute diarrhea among children. Among them, rotavirus is considered the predominant disease agent (2). Since 2006, when vaccination against rotavirus was implemented in several Latin-American countries, other enterovirus family species, such as norovirus, adenovirus, and astrovirus, have greatly increased compared to previous years (3).
The use of new technologies has strengthened diagnostic systems, leading to identifying other viral agents present in feces that were not previously identified (4-6). For example, after implementation of the rotavirus vaccine in 2006, caliciviruses were mostly begun to detect in association with gastroenteritis symptoms, followed by adenovirus, which was listed as the second or third diarrhea-causing viral agent, whereas astrovirus was predominantly observed among children up to two years of age showing severe gastrointestinal problems (2, 7, 8). These new findings reveal the lack of detailed information regarding the prevalence of enteroviruses associated with severe acute gastroenteritis. In addition, molecular detection may help identify vial co-infections in severe cases. The present study was undertaken to detect adenovirus, astrovirus, norovirus, rotavirus, and viral co-infections in stool samples of hospitalized children with acute diarrhea in the city of Chihuahua during 2010 and 2011.
2. Objectives
This study aimed to determine the co-infections of enteric viruses among children under five-years-old, hospitalized with severe acute gastroenteritis symptoms in Chihuahua, Mexico, during 2010 and 2011.
3. Methods
3.1. Stool Samples
A total of 57 samples were collected in hospitals from the city of Chihuahua, located in northern Mexico, during 2010 - 2011. Chihuahua State Children’s Hospital and General Hospital “Salvador Zubirán” for children medical attention in the city were selected in the present study for the collection of patients’ stool samples. The patients were children under five-years-old diagnosed with symptoms of severe AGE for the detection of enteric viruses of higher prevalence. Stool samples were collected in 50 mL sterile plastic cups and stored at -20°C until analysis (9).
3.2. Detection of Rotavirus Directly in Stool Samples
Diarrheal stools were analyzed using the SAS™ Rota Test Kit (SA Scientific, San Antonio, TX). The samples were thawed and 50 mg of each was re-suspended in the extraction buffer. The samples were centrifuged at 5,000 rpm for two minutes and 150 microliters were placed on the rapid strip. After one minute, the results were interpreted according to the manufacturer’s instructions.
3.3. Viral RNA and DNA Extraction
Stool sample suspensions were prepared in Phosphate Buffer Saline (PBS) at 20% wt/vol (solution added with calcium and magnesium). Total RNA was extracted by the Trizol method using 200 µL of each sample suspension (Molecular Research Center Inc., Cincinnati, OH) following the supplier’s instructions. Similarly, viral DNA was purified using 200 μL of each stool suspension, followed by treating with proteinase K (MP Biomedical, Santa Ana, CA) to reach a final concentration of 500 μg/mL. The mixtures were then incubated at 37°C for 90 min. Subsequently, 200 μL of phenol saturated with tris 2M pH 8 (Sigma-Aldrich, St. Louis, MO) was added to each mixture, vortexed for one minute, and centrifuged for 5 min at 13,000 rpm. The aqueous phase was separated and 200 μL of chloroform-isoamyl alcohol (24:1) (Sigma-Aldrich) was added. Each supernatant sample was mixed and centrifuged at 13,000 rpm for 5 min. Next, 100 μL of 7.5 M ammonium acetate (Sigma-Aldrich) was added to each supernatant; 400 μL of ice-cold 100% ethanol (CTR Scientific S.A. de C.V., Monterrey, Mexico) was added, and then the tubes were incubated overnight at -20°C. The samples were then centrifuged at 12,000 rpm for 10 min at 4°C. The supernatant was discarded and the pellet was washed three times with 500 mL of 70% ethanol, followed by centrifugation at 12,000 rpm for 10 min at 4°C. Finally, the purified RNA and DNA samples were stored at -20°C until use (9, 10).
3.4. RT-PCR and PCR
For synthesizing cDNA for Reverse Transcription (RT) amplification, 300 ng of the extracted-purified total RNA sample was used for was performed with, using Moloney Murine Leukemia Virus Reverse Transcriptase (MMLV RT) (Promega, Madison, WI), following the manufacturer’s instructions (11). For the PCR assay, 200 ng of the extracted-purified sample was amplified using previously reported primers (Table 1). Then, the amplified DNA products were analyzed to detect Rotavirus, adenovirus, astrovirus, and norovirus genotypes or the combination of them. To achieve this, BIOLASE TM DNA Polymerase was used following the manufacturer’s protocol (Bioline, Taunton, MA) (9).
| Virus/Gene | Primer | Nucleotide | References |
|---|---|---|---|
| Rotavirus | |||
| Segment 4 | Con3 | TGGCTTCGCCATTTTATAGACA | (12) |
| Segment 4 | Con2 | ATTTCGGACCATTTATAACC | (12) |
| Segment 9 | Beg9 | GGCTTTAAAAGAGAGAATTTCCGTCTGG | (13) |
| Segment 9 | End9 | GGTCACATCATACAATTCTAATCTAAG | (13) |
| Norovirus | |||
| RdRp | NV4562 | GATGCDGATTACACAGCHTGGG | (14) |
| RdRp | NV5366 | CATCATCATTTACRAATTCGG | (14) |
| RdRp | NV4611 | CWGCAGCMCTDGAAATCATGG | (15) |
| RdRp | NV5296 | CCAYCTGAACATTGRCTCTTG | (15) |
| Astrovirus | |||
| ORF2 | Mon269 | CAACTCAGGAAACAGGGTGT | (16) |
| ORF2 | Mon270 | TCAGATGCATTGTCATTGGT | (16) |
| Adenovirus | |||
| E3 | Hex1deg | GCCSCARTGGKCWTACATGCACATC | (17) |
| E3 | Hex2deg | CAGCACSCCICGRATGTCAAA | (17) |
3.5. Sequencing and Phylogenetic Analysis of Enteric Viruses
For genotyping and doing phylogenetic analysis, the PCR products were purified and amplified and the nucleotide sequence was determined by a capillary electrophoresis system (Applied Biosystems, Seoul, Korea). The sequencing analysis was performed using CLUSTALW/BioEdit sequence alignment version 7.0 and the phylogenetic tree was constructed with the MEGA version 8.0 software with 1000 bootstrapped data sets with the neighbor-joining method (18-20).
4. Results
4.1. General Clinical Characteristics of Patients
A total of 57 children were enrolled in the study. Their ages ranged from 4 to 51 months. There were 63.15% (n = 36) males and 36.85% (n = 21) females. The diarrheal episodes of the patients were up to 20 per day with a duration of one to seven days. The frequency of vomits was one to five per day and some patients had fever episodes of up to 39°C. Twenty-eight patients were positive in the rapid test for rotavirus detection.
4.2. RT-PCR
The RT-PCR results demonstrated the presence of at least one viral agent in 32 (56.14%) collected stool samples from hospitalized children in Chihuahua, Mexico. Among the enteric virus-positive samples, rotavirus was identified in 75%, adenovirus in 25%, and astrovirus and norovirus each in 6.25% of the samples. Enteric virus co-infection analysis revealed the rotavirus presence in addition to another viral agent in 14% of the analyzed samples.
4.3. Phylogenetic Tree Constructions of Enteric Viruses
After the sequencing analysis, the access numbers submitted to GenBank were assigned as MH925712 to MH925768. The access codes of control sequences were HM007189, FJ713740, HM007188, DQ235979, FN424164, KT223457, GQ129459, JN849140, GQ282613, AY 165009, EU131107, DQ779053, DQ779054, EU239216, EU033979, DQ674999, DQ078814, AB294785, AB491291, GQ303445, AB294790, AB447433, AY502023, EU310927, EF126965, EF126966, DQ078829, AY038600, AF145896, X76716, AB294779, JN167521, Y324863, JQ796915, HQ393852.2, JN203051, JQ434376, and JN799271. These sequences were used to build the phylogenetic tree.
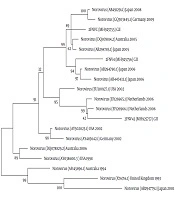
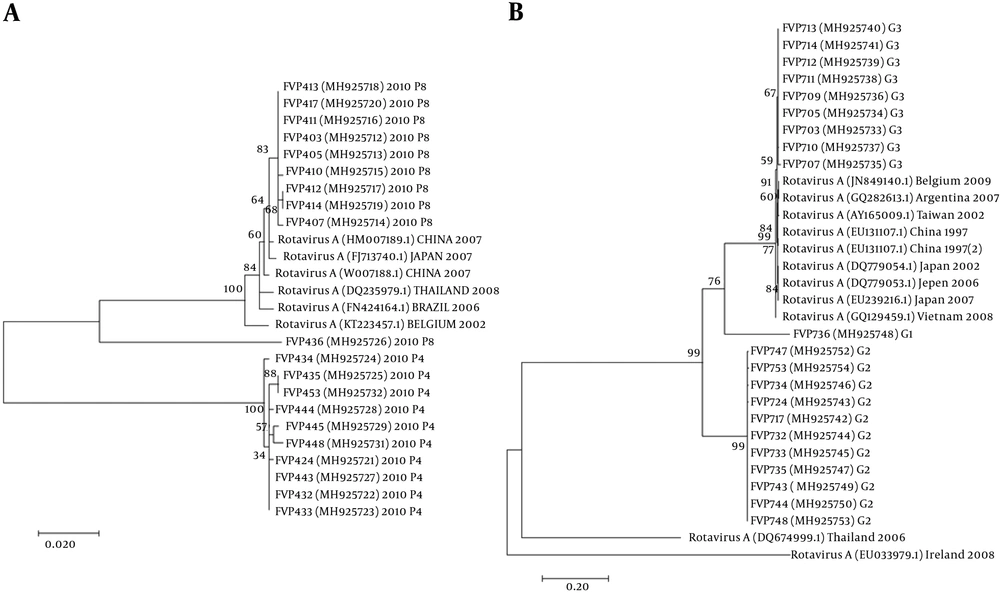
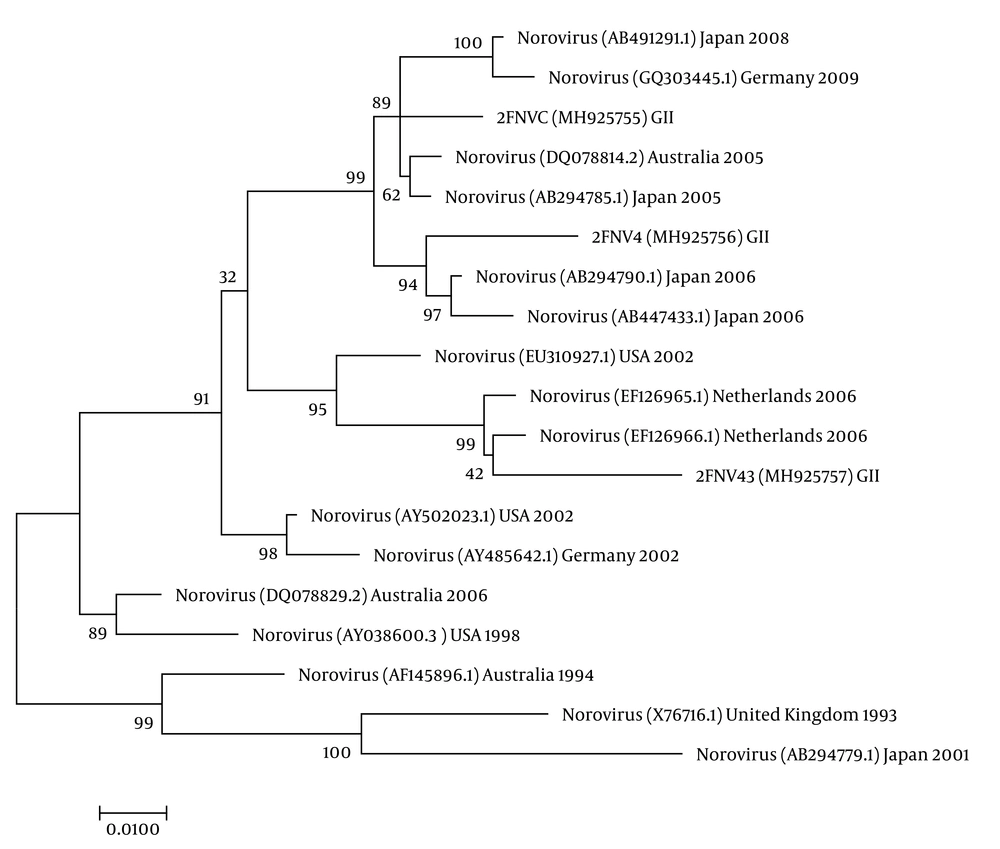
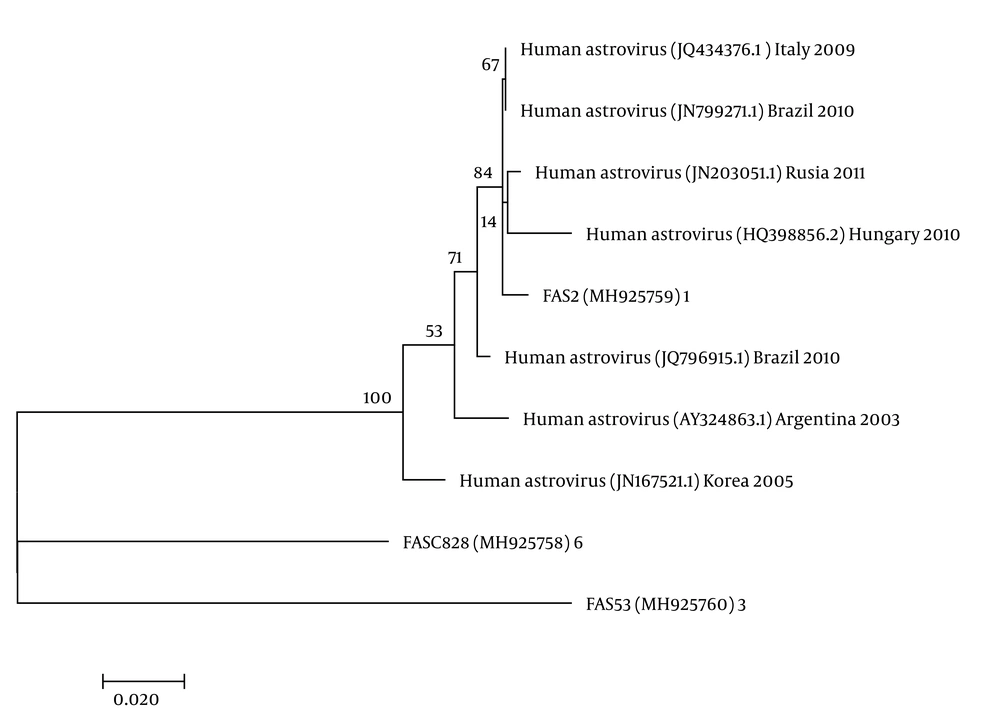
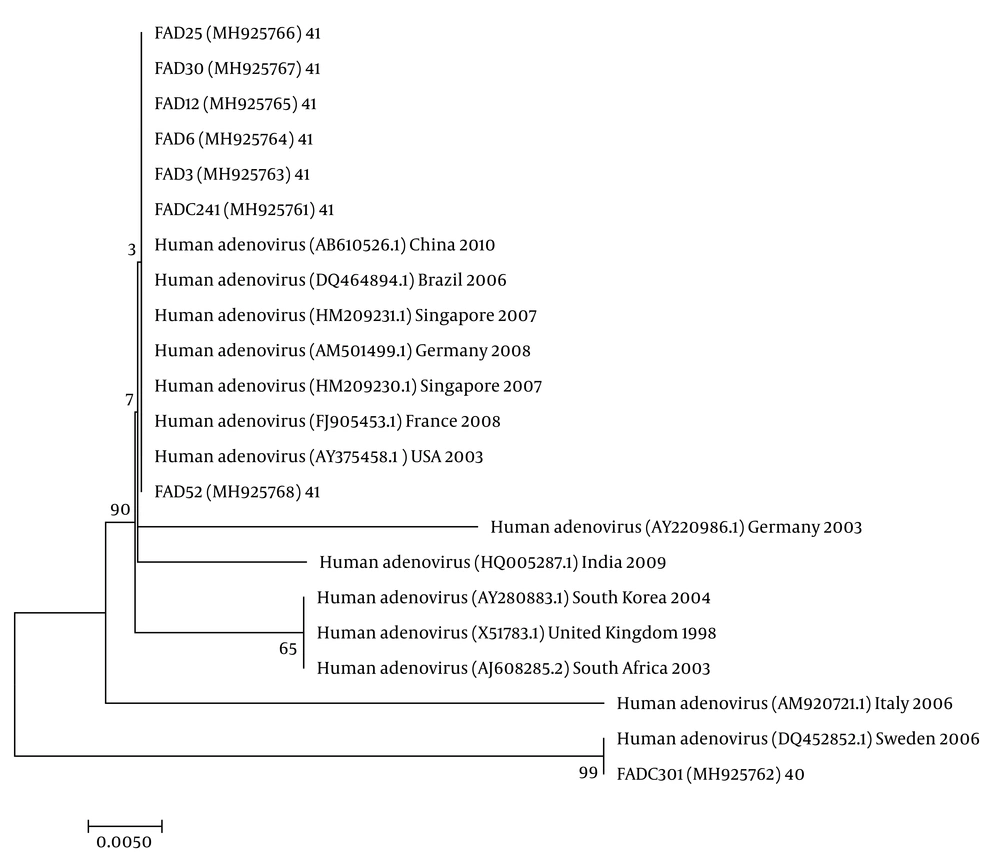
The nucleotide sequence analysis of the rotavirus VP4 genotype P[8] and genotype P[4] sequences reported in this study revealed 98-100% identity with rotavirus strain P[8] and P[4] previously reported in Belgium, Brazil, China, Japan, and Thailand (Figure 1). Moreover, the phylogenetic analysis of the VP7 genotypes showed a 98% - 99% identity with G1, G2, and G3 genotypes of rotaviruses reported in Argentina, Belgium, China, Ireland, Japan, and Taiwan (Figure 1). The analysis of norovirus sequences revealed an identity of 89% - 99% with strains of norovirus GII previously identified in Australia, Germany, Japan, the Netherlands, the United Kingdom, and the USA (Figure 2). Similarly, the astrovirus phylogenetic analysis clustered the 1 and 3 genotypes with 71% - 100% identity with astrovirus strains reported from Argentina, Brazil, Hungary, Italy, Korea, and Russia (Figure 3). Finally, the nucleotide analysis of adenovirus revealed 90% - 99% identity with the 40 and 41 genotypes from Brazil, China, France, Germany, India, Italy, Singapore, South Africa, South Korea, Sweden, United Kingdom, and USA (Figure 4).
4.4. Rotavirus Genotypes
Twenty-eight patients (n = 28) were positive in the SAS™ Rota Test, a rapid test for rotavirus detection. Out of the positive samples, 24 (85.7%) were assigned as G and P genotypes. The most prevalent combination was G2P[4] (41.7%), followed by G3P[8] (29.2%) and G1P[8] (8.3%). The G3P[4] genotype was detected in a small proportion (4.2%) of the tested samples. In addition, G1, G3, P[8], or P[4] genotype was found in 16.7% of the samples, showing mixed viral infections (Table 2).
| Strain | No. (%) |
|---|---|
| G3P[8] | 7 (29.2) |
| G2P[4] | 10 (41.7) |
| G1P[8] | 2 (8.3) |
| G3P[4] | 1 (4.2) |
| G1G3P[4]P[8] | 1 (4.2) |
| G3P[4]P[8] | 3 (12.5) |
| Total | 24 (100) |
4.5. Norovirus Genotypes
Fifty-seven patients were analyzed to identify norovirus. Norovirus was detected in two (3.5%) samples. Positive norovirus samples were assigned as the GII genotype. Fifty percent of the positive samples were in co-infection with rotavirus.
4.6. Adenovirus Genotypes
Adenovirus was tried in 57 samples of severe acute diarrhea. The virus was found in eight (14.03%) samples. Among the total positive samples, three (5.26%) co-infections were identified with rotavirus. Molecular analysis showed that the 41 genotype was identified in eight (100%) positive samples.
4.7. Astrovirus Genotypes
The molecular analysis of 57 samples showed the virus in two (3.5%) samples, of which the 1 and 3 genotypes each corresponded to 50%. Co-infection analyses revealed one co-infection with rotavirus.
5. Discussion
In the present study, the results showed the presence of rotavirus, adenovirus, norovirus, and astrovirus as causative gastroenteritis agents mainly among hospitalized children under five-years-old with a diagnosis of AGE in Chihuahua city. Enteric viruses were found in more than 50% of the samples, indicating a high incidence. Among enterovirus-positive samples, the highest incidence was related to rotavirus, followed by adenovirus, indicating differences from the incidence of enteric viruses worldwide, in which the norovirus incidence was second after that of rotavirus (21, 22). Viral co-infections were detected in 14% of the analyzed samples, where rotavirus was detected co-infecting with other detected enteroviruses (2).
The rotavirus genotyping showed that the highest prevalence was related to G3P[8], followed by G2P[4] and G1P[8], demonstrating a genotypic variation in circulating strains, as previous reports on rotavirus in the same area of this study showed the prevalence or re-emergence of the G1P[8] genotype (23, 24). In some countries, the G2P[4] genotype has shown a greater incidence than that of G1P[8] (25). Our results are similar to previous reports, which indicated variations in the prevalence of circulating rotavirus genotypes, thus demonstrating the variation of this pathogen in the children population (26). However, our results showed the prevalence of the same genotypes circulating in this area, similar to previous reports in the northeast of Mexico, as well as other countries (27). This higher prevalence of genotypes could be related to the vaccination protection, as an oscillating relationship was observed between genotypes G1P[8] and G2P[4] in previous reports (17, 28).
Norovirus was identified in 3.5% of the analyzed samples with severe AGE, whereas other reports indicated a prevalence of 5.3% in Mexico (29). Despite this, different studies showed the prevalence of this pathogen to be up to 35.2% in sporadic outbreaks of acute gastroenteritis in children under five-years-old (30). Indeed, the GII genogroup has been found in up to 92% of the analyzed samples in other countries (30-32), which represents a latent risk due to this virus incidence increase among children younger than five-years-old after the application of the rotavirus vaccine (33). Furthermore, the analysis revealed the presence of genotypes HAstV-1 and HAstV-3, indicating a high prevalence of astroviruses in this region, which is in line with reports from other countries (24, 34-37). However, the zoonotic transmission of enteric astroviruses in younger children could represent a latent risk, as previous reports identified a close relationship between AstV of humans and other animals (38).
In addition, the prevalence of Ad-41 in the analyzed samples was 25% for among the positive samples, which represents a percentage similar to that reported by other studies (39). This could be related to the viral strain present in the samples and the symptoms, since other molecular epidemiologic studies included samples without diarrheic symptomatology (40-42). After the introduction of the rotavirus vaccine and advances in diagnostic techniques, several studies showed an increase in enteric virus co-infections. In the present study, adenovirus, astrovirus, and norovirus co-infections with rotavirus were determined. Our results indicated that, unlike other studies, only was rotavirus found co-infecting with other enteroviruses, where no co-infection between adenovirus, astrovirus, and norovirus was detected, whereas reports from South America indicated viral co-infections in samples from patients diagnosed with AGE (2, 43, 44). In summary, co-infections between enteric viruses are increasing, which represents a serious problem for human health due to the emergence of these viruses in children under five-years-old (2, 27, 43-45).
5.1. Conclusions
Our data support the need for constant monitoring of viral enteropathogens, due to the global epidemiological variations after rotavirus vaccination, as well as the molecular characterization of circulating viruses associated with acute diarrheal diseases.