1. Background
Cryptosporidium is recognized as an important and common cause of diarrhea in both animals and humans (1). In immunocompetent hosts, it may produce self-limiting acute diarrheal disease. Young animals and children under five years of age are most often infected with this genus. In immunocompromised patients, it may also cause a prolonged severe disease. This coccidium is a major opportunistic pathogen in AIDS patients (2-12). Cryptosporidium oocysts in fecal samples are characterized via microscopic examination after concentrating using different methods, including flotation and formol-ether concentration techniques. To characterize these species, new tools have been introduced in molecular biology (13). Of the 31 Cryptosporidium species that have been recognized as valid, more than 20 species and genotypes are identified in humans, including Cryptosporidium hominis, C. parvum, C. meleagridis, C. felis, C. canis, C. cuniculus, C. ubiquitum, C. viatorum, C. muris, C. suis, C. fayeri, C. andersoni, C. bovis, C. scrofarum, C. tyzzeri, C. erinacei, and Cryptosporidium horse, skunk, and chipmunk I genotypes, with C. hominis and C. parvum being the most commonly reported ones (14, 15).
2. Objectives
3. Methods
3.1. Study Population
This study was performed from August 2016 to August 2018 among 764 children with the acute diarrheal disease under 10 years of age, who were admitted to Aliasghar Pediatric Hospital and the Reference Laboratory of Zahedan, Sistan and Baluchestan, Iran. Children attending these hospitals came from different urban and rural areas of the province and they all resided in poor regions. An evaluation was made on each patient at the time of sample collection based on the information provided by the children’s mothers and a review of the patients’ medical histories. The inclusion criteria included an age of ≤ 10 years and no history of antiparasitic drug use during two weeks before the test. The exclusion criteria included children suffering from gastrointestinal disorders including diarrhea, stomach pain, bloating, and nausea or vomiting.
3.2. Fecal Samples
The fecal samples were collected in cardboard boxes within two days of hospital admission or when the child presented as an outpatient. A stool container was given to the mother and she was asked to return a stool sample on the next day. The specimens were then transferred to our laboratory and concentrated using a water-ether concentration procedure.
3.3. Microscopic Examination
For identifying and recovering Cryptosporidium oocysts, the water-ether concentrated stool samples were microscopically examined at 1000X magnification after modified acid-fast staining (21-23).
3.4. DNA Extraction
For DNA extraction, a FavorPrep® Stool DNA Extraction Kit (Favorgen, Taiwan) was used based on the manufacturer’s protocols.
3.5. PCR Amplification
For the amplification of the 18S rRNA gene, a nested PCR protocol was applied, as previously described (24). A C. parvum-positive control was also included to validate the results. The positive sample for C. parvum (previously identified by PCR-RFLP) used in the assays was a gift from Dr. Hadi Mirahmadi, ZAUMS, Zahedan, Iran.
3.6. RFLP Assay
For RFLP analysis, the secondary PCR products were digested with restriction enzyme Vsp1. Horizontal electrophoresis was performed to separate the restriction fragments in the 2% agarose gel.
3.7. Sequence Analysis
A purification kit was used to purify the secondary PCR amplicons (Bioneer, Daejeon, South Korea). An automated DNA analyzer was also used for sequencing in both directions (ABI 3730 XL, Bioneer, South Korea). The sequences were edited manually in BioEdit (http://www.mbio.ncsu.edu/BioEdit/page2.html) and aligned with reference sequences for genotype identification in BLASTN software (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The multiple-sequence alignment was carried out and a phylogenetic tree was plotted using Clustal W and neighbor-joining (NJ) method based on the Kimura two-parameter model in MEGA version 6.0 (25). The bootstrap method was used to evaluate the reliability of the NJ tree with 1000 replicates.
4. Results
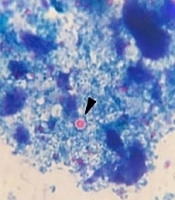
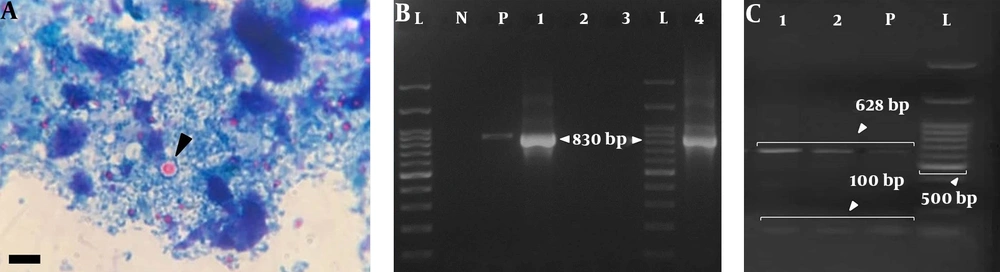
Of 764 stool samples, 0.91% (7/764) were positive for Cryptosporidium oocysts presenting as particles in the microscopic examination (Figure 1A). Overall, 28.6% (2/7) of the positive samples were successfully DNA amplified via nested PCR. The 830-bp products obtained from the secondary PCR were in accordance with the positive control sample (Figure 1B). The expected product was not produced by negative controls based on the nested PCR. No amplification was observed for five microscopically positive fecal samples.
A, Cryptosporidium spp. oocyst-like particles detected in stool specimens of symptomatic patients in the current study (scale bar, 10 μm); B, nested-PCR products on the ethidium bromide-stained 2% agarose gel. Lanes 1 and 4: C. parvum-positive samples (830 bp fragment); lanes 2 and 3: negative samples; lane N: negative control; lane P: positive control; lane L: 100-bp DNA ladder (Cinnagen/Iran); C, gel electrophoresis of DNA fragments obtained after digestion with Vsp1 restriction enzyme. Lanes 1 and 2 (positive samples) and P (positive control) show the banding patterns after digestion with Vsp1 (GenBank Accession no. MK530165, MK530166, and MK530167); lane L is a 100-bp DNA ladder.
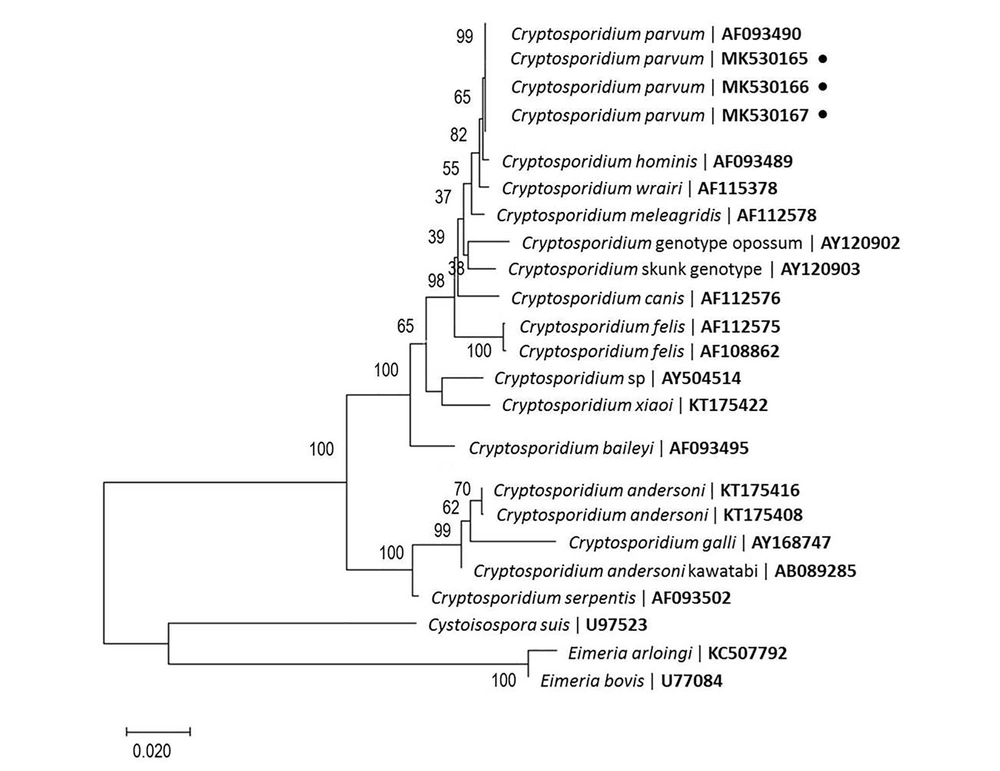
The secondary PCR products, which were exposed to Vsp1 restriction enzyme digestion, indicated the presence of C. parvum in the positive control and fecal samples, as gel electrophoresis indicated two bands with sizes of 100 bp and 628 bp (Figure 1C). The PCR-positive fecal and positive-control samples were successfully sequenced. In total, 23 nucleotide sequences were included in the analysis (Figure 2). According to the search of 18S rRNA sequences (560 bp - 815 bp) against the published sequences of other Cryptosporidium species, the greatest homology was attributed to C. parvum (99% - 100%). The nucleotide sequences in our study were deposited in GenBank (accession No., MK530165 and MK530166). Moreover, the nucleotide sequence of the positive control sample was deposited in GenBank (No., MK530167).
Phylogenetic tree based on 18S sequences, constructed according to the NJ method, showing the position of Cryptosporidium species. Cystoisospora suis and Eimeria spp. were used as outgroups. The percentage of replicate tree in which the associated taxa were clustered together in the bootstrap test (1,000 replicates) is shown next to the branches.
5. Discussion
The present study showed that C. parvum is an important cause of diarrhea in children younger than 10 years in Zahedan, Iran. This finding was not unexpected, as intestinal parasites are common in this area and the studied population represented the lowest income class in the region, with poor environmental sanitation facilitating frequent exposure to enteropathogens. The prevalence of cryptosporidiosis has been examined in diarrhea children in different areas of Iran. In studies by Keshavarz et al. (17) and Saneian et al. (18), low prevalence rates were reported in the central regions of Iran (2.5% and 4.6%, respectively). On the other hand, Mirzaei (19) reported a high prevalence rate in the southern regions of Iran (35%). In a study by Dabirzadeh et al. (20) from Zabol, microscopic examinations indicated the rate of positive samples as 9.5% (19/200). The findings represent geographical differences in the prevalence of Cryptosporidium in Iran.
Microscopic examination is commonly used to detect Cryptosporidium oocysts in fecal samples after concentrating using different methods, including flotation and formol-ether concentration techniques. The efficacy of molecular methods has been confirmed for the identification and classification of Cryptosporidium oocysts. In the present study, nested-PCR and RFLP assays were conducted to identify Cryptosporidium species in stool samples. The rate of positivity was lower in PCR than in microscopic examination. Previous studies also reported similar findings, which might be attributed to DNA inadequacy due to the low number of oocysts in stool samples (26-28).
Cryptosporidium parvum and C. hominis are usually responsible for the disease in humans. Although other animals, especially cattle, and humans are the reservoir hosts for C. parvum, humans are the only reservoirs for C. hominis. In addition, the distribution of these infective parasitic species varies in different countries. A previous study showed that C. parvum accounted for most cases of the disease among humans in the United Kingdom (61.5%) (29). In addition, different seasonal patterns were reported for these species. The peak of C. parvum infection occurs in the late spring while the peak of C. hominis infection is in the fall (30). The present study was conducted from August 2016 to August 2018. Evidence suggests that C. hominis accounts for most human infections in the United States, Kenya, Guatemala, Australia, and Peru (31-33). In Iran and its neighboring countries, C. parvum is more frequently reported than C. hominis (33-36). Cryptosporidium was identified in 4 out of 707 school children from Turkey (0.6%); all the isolates belonged to the C. parvum bovine genotype (34). The current results are consistent with the reports from Saudi Arabia, Iran, Kuwait, and Turkey (8, 22-36).
Cattle are a major source of zoonotic cryptosporidiosis with C. parvum. Some of the transmission routes include direct contact with infected calves and water contamination with the cattle manure (37). According to a survey from Northwestern regions of Iran, C. parvum and C. andersoni infected 10.5% of the cattle (38). However, the transmission route cannot be achieved by the determination of species. Some of the C. parvum subtypes are found only in humans, indicating that some of these subtypes have anthroponotic transmission and some others are zoonotic (39).
5.1. Conclusions
We evaluated the distribution of cryptosporidial infection in children (< 10-years-old) in Zahedan, Iran. The presence of this infection in children can be threatening to other susceptible people in this region. Nevertheless, it is important to conduct further epidemiological studies to evaluate the risk of cryptosporidial infection in other susceptible people, including immunocompromised patients.