1. Background
Mycobacterium tuberculosis is a pathogen that causes tuberculosis. It can invade all organs of infected patients, but lung infection is most common. Tuberculosis is a chronic respiratory infection. Before the application of anti-tuberculosis drugs, tuberculosis was known as the “white plague” and its high morbidity and high mortality seriously threatened human health. The application of anti-tuberculosis drugs has effectively controlled tuberculosis and has been promising in curing tuberculosis. The emergence of Drug-resistant (DR) and Multidrug-resistant (MDR) strains has led to complications and difficulty in the treatment of tuberculosis (1). Early detection of these strains before the initiation of treatment is important for implementing an effective treatment regimen to prevent the spread of these strains, thus avoiding the further spread of DR tuberculosis. China is one of the 14 countries in the list of three high-burden countries for tuberculosis, tuberculosis/HIV, and MDR tuberculosis as identified by the World Health Organization (WHO) during 2016 - 2020 (2).
Multidrug-resistant tuberculosis is defined as resistance to at least two first-line drugs rifampicin (RIF) and isoniazid (INH). Mycobacterium tuberculosis resistance is mainly caused by drug activating enzymes or drug target mutations (3). Until today, INH is still the cornerstone of modern tuberculosis chemotherapy. However, M. tuberculosis is becoming more resistant to INH (4). A variety of genes are involved in M. tuberculosis resistance to INH. It is reported that frequent mutations primarily focus on the katG and inhA regulatory regions while very few mutations occur in the ahpC gene (5). KatG has a length of 2223 bp and synthesizes a catalase-peroxidase enzyme, which is involved in the synthesis of mycolic acid (6) while inhA is involved in the synthesis of fatty acids (7).
Tuberculosis is a chronic disease with a long treatment cycle. Before anti-infective treatment, the sensitivity of antibacterial drugs should be clear, which is the key to successful treatment. Molecular biology is a common method to detect drug resistance genes of M. tuberculosis. To understand the drug resistance characteristics of M. tuberculosis in Shanghai and its main drug resistance mechanism, this study used the clinical isolates of M. tuberculosis in Shanghai as experimental strains.
2. Objectives
The study determined the frequency of mutations in three specific resistance genes (katG, inhA, and ahpC) in M. tuberculosis strains and assessed the correlation between the minimum inhibitory concentration (MIC) of antibacterial drugs and different mutation sites.
3. Methods
3.1. Clinical Isolates
We collected 92 INH-resistant and 30 INH-susceptible clinical isolates of M. tuberculosis from the library of strains in Shanghai Pulmonary Hospital. All 122 strains of M. tuberculosis were isolated from patients with pulmonary tuberculosis during the period between January and June 2018. Each strain corresponded to a single patient. We identified all strains as M. tuberculosis using the mycobacteria growth indicator tube (MGIT) liquid culture method (BD Biosciences, New Jersey, USA) and MPT-64 antigen detection. The 16SrRNA sequence was then amplified to confirm the previous identification results. All strains were cultured on Lowenstein-Jensen (LJ) and MGIT liquid media for subsequent experiments.
3.2. Phenotypic Drug Susceptibility Testing
Cultures on the LJ medium were collected and tested for drug susceptibility to first-line drugs, RIF, ethambutol (EMB), and INH, as well as second-line drugs, ofloxacin (LFX), moxifloxacin (MOX), amikacin (AMK), kanamycin (KAN), ethionamide (ETH), and cycloserine (CYC). Drug susceptibility testing was performed using the broth microdilution method and Roch broth containing OADC (oleic albumin dextrose catalase) supplement (Thermo, USA). The critical concentrations of these drugs were as follows: 1 µg/mL for RIF, 5.0 µg/mL for EMB, 0.2 µg/mL for INH, 2.0 µg/mL for OFX, 0.5 µg/mL for MXF, 4.0 µg/mL for AMK, 5.0 µg/mL for KAN, 5.0 µg/mL for ETH, and 25.0 µg/mL for CYC. If MICs of INH were > 0.2 µg/mL, the strain was considered resistant according to the WHO recommendations. Pan-susceptible M. tuberculosis strain H37Rv (ATCC27294) was used as a reference.
3.3. Mycobacterium tuberculosis DNA Isolation
The frozen isolates were subcultured on the LJ medium for four weeks. The crushed bacterial colonies were recovered in 200 μL of 1 × TE (Tris-EDTA) buffer and boiled at 100°C for 10 minutes to inactivate bacteria and release the mycobacterial DNA. The recovered DNA was immediately used for PCR amplification stored at -20°C until use. The M. tuberculosis reference strain H37Rv was used as the control.
3.4. PCR Amplification and Sequencing of katG, inhA, and ahpC
The amplification of drug-resistant genes included an amplicon of 820 bp for the katG gene amplified using the forward primer (5′CGGCGATGAGCGTTACAGC3′) and reverse primer (5′TCGTTGACCTCCCACCCGACTTG3′). For the inhA gene, an amplicon of 650 bp was amplified using the forward primer (5′AGGTCGCCGGGGTGGTCAGC3′) and reverse primer (5′ATGTTGATCAGGGTCTGC3′). For the ahpC gene, an amplicon of 730 bp was amplified using the forward primer (5'GCAACCAAATGCATTGTCCG3') and reverse primer (5'GAGCTTTTCTATACTCATTG3').
The PCR mixture was prepared in a volume of 50 µL as follows: 25 µL 2 × PCR Mixture (Sangon, Shanghai, China), 2 µL of DNA template, and 2 µL of DNA template. The reaction was carried out in a thermal cycler, as follows: 94°C for 5 minutes, followed by 35 cycles of denaturation at 94°C for 30 seconds, primer annealing (katG at 61°C, inhA at 63°C, and ahpC at 53°C) for 30 seconds, extension at 72°C for one minute, and final elongation at 72°C for 10 minutes. The PCR products were separated by 1% agarose gel electrophoresis. A positive product of the target gene fragment was sent to the company (Tsingke, China) for sequencing. Resulting sequences were compared with wild-type sequences of M. tuberculosis H37Rv using Bio-Edit software (version 7.0.9).
3.5. Statistical Analysis
All statistical analyses were performed using SPSS (V 20.0; SPSS Inc.). Data were compared by the χ2 test. For all analyses, a P value of < 0.05 was considered statistically significant.
4. Results
4.1. Drug Resistance Profiles of Isoniazid-Resistant Mycobacterium tuberculosis Clinical Isolates
Based on drug susceptibility testing results, 73 (79.3%) isolates were resistant to RIF and 30 (32.6%) were resistant to EMB. There were 73 (79.3%) MDR isolates and 19 (20.7%) non-MDR INH-resistant isolates. The drug susceptibility testing results of 92 INH-resistant isolates against six second-line anti-tuberculosis drugs showed that 39 (42.4%) isolates were resistant to LFX and MOX, 17 (18.5%) isolates to AMK, 19 (20.7%) isolates to KAN, 12 (13.0%) isolates to ETH and eight (8.7%) isolates were resistant to CYC (Table 1). Among 92 INH-resistant clinical isolates, 15 strains were found to have MICs of < 1 µg/mL and 77 had MICs of ≥ 1 µg/mL (Table 2).
| Drug | No. (%) |
|---|---|
| RIF | 73 (79.3) |
| EMB | 30 (32.6) |
| LFX | 39 (42.4) |
| MOX | 39 (42.4) |
| AMK | 17 (18.5) |
| KAN | 19 (20.7) |
| ETH | 12 (13.0) |
| CYC | 8 (8.7) |
| Total | 92 |
| MDR | 73 (79.3) |
| non-MDR INH-resistant | 19 (20.7) |
Abbreviations: AMK, amikacin; CYC, ethionamide; EMB, ethambutol; ETH, ethionamide; KAN, kanamycin; MDR: rifampicin and isoniazid-resistant; MOX, moxifloxacin; non-MDR INH-resistant: non-multidrug resistant but INH-resistant; OFX, levofloxacin; RIF, rifampicin.
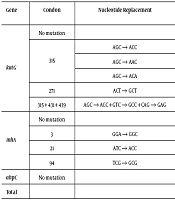
| Gene | Condon | Nucleotide Replacement | Amino Acid Replacement | No. (%) | MICs (µg/mL) (No.) |
|---|---|---|---|---|---|
| katG | No mutation | 28 (30.4) | < 1 (10), ≥ 1 (18) | ||
| 315 | AGC → ACC | Ser315 → Thr | 58 (63.0) | < 1 (5) , ≥ 1 (53) | |
| AGC → AAC | Ser315 → Asn | 2 (2.2) | 1 (1), > 4 (1) | ||
| AGC → ACA | Ser315 → Thr | 2 (2.2) | 4 (1), 2 (1) | ||
| 271 | ACT → GCT | Thr271 → Ala | 1 (1.1) | > 4 (1) | |
| 315 + 431 + 439 | AGC → ACC + GTC → GCC + CAG → GAG | Ser315 → Thy + Val431 → Ala + Gln439 → Glt | 1 (1.1) | > 4 (1) | |
| inhA | No mutation | 87 (94.6) | |||
| 3 | GGA → GGC | Synonymous mutation | 1 (1.1) | 0.5 (1) | |
| 21 | ATC → ACC | Ile → Thr | 3 (3.3) | 1 (1), 2 (2) | |
| 94 | TCG → GCG | Ser → Ala | 1 (1.1) | 0.5(1) | |
| ahpC | No mutation | 92 | |||
| Total | 92 |
4.2. Mutations in the katG, inhA, and ahpC Genes
Out of 92 INH-resistant clinical isolates, 64 (69.6%) isolates had an amino acid substitution in katG, with a vast majority carrying Ser to Thr (AGC → ACC) substitution at codon 315. The mutation at codon 315 prevailed in the katG gene with three mutations, including Ser315 → Thr (AGC → ACC) (63.0%), Ser315 → Asn (AGC → ACC) (2.2%), and Ser315 → Thr (AGC → ACA) (2.2%). Among these isolates, one (1.1%) isolate showed simultaneous mutations at codons 315, 431, and 439 (1.1%). Furthermore, a single mutation at codon 271 was observed in one (1.1%) isolate. Mutations in the inhA regulatory region were observed in five (5.4%) INH-resistant clinical isolates and a single mutation at codons 3, 21, and 94 was observed in one (1.1%) isolate, three (3.3%) isolates, and one (1.1%) isolate, respectively (Table 2).
No mutation was observed in the ahpC gene. Only had one (1.1%) isolate mutations in both katG and inhA. Of 30 INH-susceptible clinical isolates, only had one (3.3%) isolate an amino acid substitution Ser315 → Thr (AGC → ACC) in katG (P < 0.05) (Table 3). However, 30 INH-susceptible isolates showed no mutation in the inhA and ahpC genes. Out of 77 high-MICs (MIC ≥ 1 µg/mL) INH-resistant clinical isolates, 53 (68.8%) isolates were associated with Ser315 → Thr (AGC → ACC) substitution while only five (33.3%) isolates had this mutation out of 15 isolates with low MICs (MIC < 1 µg/mL) (P < 0.05) (Table 3). In addition, the frequency of Ser315 → Thr (AGC → ACC) substitution in the katG gene was more in MDR isolates (67.1%) than in non-MDR INH-resistant isolates (47.4%) (P < 0.05) (Table 3).
| Phenotypic DST | Ser315 → Thr (AGC → ACC) Substitution in katG | Total | |
|---|---|---|---|
| Mutation | No Mutation | ||
| INH-susceptible | 1 | 29 | 30 |
| INH-resistant | 58 | 34 | 92 |
| Total | 59 | 63 | 112 |
| Non-MDR INH- resistant | 9 | 10 | 19 |
| MDR | 49 | 24 | 73 |
| Total | 58 | 34 | 92 |
| Lower MICs | 5 | 10 | 15 |
| High MICs | 53 | 24 | 77 |
| Total | 58 | 34 | 92 |
5. Discussion
Anti-tuberculosis drug resistance poses a major threat to human health. It is usually caused by a change in the drug target due to a mutation in the chromosomal gene of M. tuberculosis. Information on the genetic diversity of M. tuberculosis plays an important role in controlling tuberculosis (8). This can help us monitor the disease, determine the origin and spread of pathogens in the area, and effectively prevent and control the disease. Mycobacterium tuberculosis resistance to INH is associated with mutations in several genes. Isoniazid is known to cause the exclusive lethal action to M. tuberculosis cells because of the pathogen’s own catalase-peroxidase (katG) enzyme that converts INH to a very reactive radical. Isoniazid is a pro-drug and its activation in a cell is performed by catalase-peroxidase, coded by the katG gene in M. tuberculosis (9). Mutations in the inhA regulatory region are known to induce overexpression of inhA and promote INH resistance by increasing the number of target molecules (7, 10). Mutations in the katG and inhA genes are most clinically relevant and determine resistance in most clinical isolates (11, 12).
In our study, 64 (69.6%) isolates had an amino acid substitution in katG. The prevalence of mutations in katG varies strongly among different regions of the world. The mutation rate was 98% in Russia (13), 66.7% in Finland (14), 60% in South Africa (15) and other areas of China (16), and 46% in Switzerland (17). Our results were similar to results from Finland, South Africa, and other areas of China, but lower than the result from Russia and higher than the result from Switzerland. The differences in clinical medication habits in different countries and limitations of selected experimental strains may be the main reasons for the inconsistent incidence of katG gene mutations. Of particular interest, a vast majority of isolates showed a Ser to Thr (AGC → ACC) substitution at codon 315 (63.0%). This is consistent with the results reported previously (18, 19). We also detected mutations at codons 315, 431, and 439 of katG in one isolate, meanwhile, the Thr271 → Ala (ACT → GCT) substitution was detected in another isolate.
These results have not been previously reported. We detected two strains with a high level of resistance to INH (MIC > 4 µg/mL). Therefore, their possible participation in the process of resistance to INH needs to be further explored. Mutations in the inhA regulatory region were observed in five (5.4%) INH-resistant isolates, which was lower than the result recorded in Kazakhstan (20). We only detected a single mutation at codon 3 in one (1.1%) isolate, codon 21 in three (3.3%) isolates, and codon 94 in one (1.1%) isolate and their amino acid substitutions were synonymous mutation (GGA → GGC), Ile21 → Thr (ATC → ACC), and Ser94 → Ala (TCG → GCG), respectively. It is worth noting that we did not detect the common frequent mutation (C-15T) in the inhA gene while this mutation was reported in phenotypically resistant isolates in many countries such as Myanmar, the Kyrgyz Republic, and Ecuador (18, 19, 21). It might be related to regional differences. In our study, no mutation was observed in the ahpC gene, which might be related to the sample size of this study that needs to be expanded for further studies. Only have one (1.1%) isolate mutations in both katG and inhA. This double mutation also occurred with lower frequency in our study that in studies from the Kyrgyz Republic and Kazakhstan (19, 20).
We also observed a strong correlation between the MICs of INH and the mutation of Ser315 → Thr (AGC → ACC) in our isolates. Out of 77 high MIC (MIC ≥ 1 µg/mL) INH-resistant clinical isolates, 53 (68.8%) isolates were associated with Ser315 → Thr (AGC → ACC) substitution while only had five (33.3%) isolates this mutation out of 15 isolates with low MICs (MIC < 1 µg/mL). These findings are in agreement with previous studies (22). In addition, the frequency of Ser315 → Thr (AGC → ACC) substitution in the katG gene was more in MDR isolates (67.1%) than in non-MDR INH-resistant isolates (47.4%). Therefore, the most common mutation was Ser315 → Thr (AGC → ACC) (63.0%) in the isolates of our study, and most of them showed high-level resistance to INH (MIC ≥ 1 µg/mL). These results indicate the Ser315 → Thr (AGC → ACC) substitution was likely associated with MDR and high-level resistance to INH in our study.
5.1. Conclusions
In conclusion, our results further enhance our understanding of the molecular mechanisms involved in INH resistance. By detecting frequent mutation sites in the katG and inhA genes and discovering new mutation sites, we provided a basis for further study of the mechanism of INH resistance and rapid detection of DR-Tuberculosis.