1. Background
Hepatitis C virus (HCV) is an RNA virus from the Flaviviridae family, which currently has contaminated about 3% of the world's population, which is more than 170 million people (1). Infection with this virus stimulates a wide range of acquired responses. The treatment of HCV contains a combination of pegylated interferon alpha (PEG-INF) and ribavirin, leading to a reduction in the variability in viral replication and clinical outcomes. The treatment response is defined as a sustained viral response (SVR), a negative PCR test conducted six months after the end of the treatment. The rate of persistent viral response among individuals infected with genotype 1 of hepatitis C virus is about 40 - 50% after 12 months of treatment (2, 3). In addition to both innate and host-acquired immunity systems which play essential roles in facing with the infection with hepatitis C, other factors such as genotype of the virus, transmission method, and genetic content of individuals contribute to the susceptibility of people to hepatitis C virus. Therefore, many studies have been conducted based on the potential genetic predisposing factors involved in response to the treatment and the natural trend of the hepatitis C virus (4, 5).
Among the genes that play the most crucial role is the Interleukin 28B gene (IFNL3) encoded IFNλ3, which is from type III interferons (6). IL 28B provides an antiviral state and increases the resistance of other cells to viral infection. Also, IL 28B can increase the expression of interferon gamma, MX' gene, 5'2'-oligo-adenylate synthase, and the cytotoxicity of TCD8 + cells. Similar to other type-III interferons, the IL28B signaling pathway is via IL 10R and IL 28R subunits and STAT-JAK pathway. Interferon λ receptors exist on many cells, including the liver and epithelial cells, and play an important role in liver function (7, 8). IFNL3 gene is located on the long arm of human chromosome 19 (9). The most important polymorphisms, known in the IFNL3 gene, are rs-12979860 and rs-8099917 variations (10). They are single-nucleotide polymorphisms located respectively at 3 kb and 8 kb both on the upstream of the IFNL3 gene (11, 12). Studies show that the allele C of the rs-12979860 polymorphism is strongly associated with continuous and consistent response to anti-HCV drug therapy and relationship to spontaneous purification (13, 14).
The optimal genotype (CC) of the rs-12979860 variant increases the probability of a susceptible viral response in comparison with the undesirable genotype (TT) that prevails in Hispanic people two times and in African Americans three times higher than European people.IFNL3 gene polymorphism likely affects the susceptibility of viral response relative to the human race (15). The C allele auto-eliminates the HCV infection, which researchers have referred to as a protective allele. The genome-wide association study (GWAS) has shown that IFNL3 gene polymorphism can be a prognostic factor in response to treatment in patients with hepatitis C virus (16).
The effect of these polymorphisms was different in various races in several studies, so that in African subjects, there was a lower response in comparison to European subjects (17). Besides, various studies showed a relationship between the IFNL3 gene and the rate of virus amplification in the early days of infection with the virus (18). The prevalence of this disease varied in different parts of Iran. In the study which was carried out in Isfahan, TT, CT and CC genotypes were 17%, 41% and 42%, respectively (19). However, this ratio in the studied patients in Yazd showed that the highest level of IL-28 gene genotype in HCV patients was related to CT genotype (55.4%) (20). As well, the prevalence of CC, CT, and TT genotypes in the study in Mazandaran was 41.4, 41.6, and 17.3%, respectively (21).
2. Objectives
Due to the high diversity of the IFNL3 gene and the appropriate anti-HCV drug therapy use, we decided to investigate its polymorphism in the HCV patient's population for the first time in Tehran province, Iran.
3. Methods
3.1. Inclusion and Exclusion Criteria
This case-control study was conducted between 2017 and 2018. One hundred and twenty patients with hepatitis C admitted to the Genetic Diagnostic Laboratory Center were categorized and evaluated in case subjects. Also, we considered 120 people as healthy controls in this study and entered the two groups after obtaining their consent. Patients were - HIV and - hepatitis B virus (HBV). We received their information from their relevant file. Then, we took six cc of blood and serum from every patient and healthy subject and infused them into an anticoagulant tube coated with K2 EDTA (AVAPEZESHK). Next, we conducted the PCR according to the instructions kit to extract the RNA virus from the collected serum via the NucleoSpin® RNA Virus Kit (Macherey-Nagel) and verify the positive or negative HCV. After that, we performed the HCV genotyping test to determine the genotypes for HCV PCR+ patients.
3.2. Extracting Genomic DNA
We extracted the genomic DNA from the Genomic DNA Miniprep Kit (Cat. No: BS88503) according to its current instructions. Then, we kept the samples in the freezer.
3.3. Determination of IFNL3 Gene Polymorphism Genotype by PCR
We used the PCR-RFLP technique to determine the polymorphism genotype of rs12979860. Also, we designed two Forward and Reverse primers reproducing a 242bp fragment (Table 1). The PCR reaction was carried out at a final volume of 25 μL with a concentration of 1 μM for each of the Forward and Reverse primers, 2 M concentration for MgCl2, 0.5 mM concentration of dNTP, 2.5 μL PCR buffer, and 0.5 μL of SmarTaq DNA polymerase enzyme (Sinaclon, Iran). To each micro-tube PCR, 1.5 μL of genomic DNA samples were added at a concentration of 100 - 200 ng, and the PCR program was performed on the following conditions in the ABI apparatus.
Primary denature for 5 minutes at 94°C followed by 38 cycles of incubation for 30 seconds at 94°C. Connecting the primer to the genome for 30 seconds at 66°C and expanding for 20 seconds at the temperature of 72°C and final expansion for 15 minutes at 72°C. After completion of the PCR, its products were electrophoresed on a 1% agarose gel for 20 minutes. Subsequently, the 242-bp band was observed on the gel. Then, the products were digested into smaller pieces to determine the genotype of rs1297986 polymorphisms in the IFNL3 gene by the BstUI restriction enzyme. The enzymatic reaction mixture contained 1 μL of buffer and 1 μL of the enzyme added to the PCR product. The samples were then placed at 37°C for 48 hours and finally electrophoresed on a 3% agarose gel for 20 minutes at 90 volts.
| Name | Sequence (3’→ 5’) | The Number of Nucleotides/TM | Restriction Enzyme |
|---|---|---|---|
| rs12979860 [C/T] F | GCTTATCGCATACGGCTAGG | 20 - 59.6 C | BstUI |
| rs12979860 [C/T] R | AGGCTCAGGGTCAATCACAG | 20 - 60.7 C | BstUI |
3.4. Statistical Analysis
Data were collected from patients and statistical analysis was performed via SPSS version 17. ANOVA test was used for analyzing the differences between 3 genotypes and the significance level was statistically significant (P < 0.05).
4. Results
4.1. Prevalence and Frequency of IFNL3 Gene Alleles
The findings of the present study showed that among the 120 patients, 30 (25%) were female, and 90 (75%) were male, whereas in the healthy group, 42 (35%) were female and 78 (65%) were male. The mean age of the patients was 35.03 (20 - 78 years old), and the mean age of the control group was 32.3 (16 - 70 years old). There was a significant difference between the two healthy and HCV patients regarding gender (P < 0.02). The results showed that the frequency of CC in healthy women was 66.7% and 47.6% in the female patients, while TT frequency was 20% in women with HCV and 9.5% in healthy women and 13.3% in CT in healthy women and 42.9% Which is statistically significant (P = 0.02331). While the frequency of CC in healthy men was 44.3% and 25.6% in male patients, the prevalence of TT in healthy men was 15.9% and in male patients, 30.8%.
The frequency of CT in healthy men was 39.8% and 43.6% in male patients with a statistically-significant difference in the results (P = 0.0166). In conclusion, the frequency of C and T alleles in patients was 132 and 108 (55% and 45%), respectively, whereas, this rate was 160 and 80 (66.6% and 33.4%) in healthy subjects for C and T. In general, the risk of developing a virus in the CC population was shown to be 2,000 times lower than that of other people who are TT and TC, which is significant (Table 2). In addition, the risk of developing the HCV virus in a TT genotype person is 0.671 times higher than people with CC and TC. Moreover, the risk of developing the virus in people with CT genotype is 0.646 times higher than those with CC and TT.
| CC vs. TT + TC | |||||
|---|---|---|---|---|---|
| P | OR | 95% CI for OR | |||
| Lower | Upper | ||||
| Total | IFNL3 [1] | 0.009 | 2.000 | 1.185 | 3.377 |
| Male | IFNL3 [1] | 0.013 | 2.308 | 1.193 | 4.464 |
| Female | IFNL3 [1] | 0.112 | 2.200 | 0.833 | 5.809 |
4.2. The Frequency of Polymorphism Genotypes rs12979860 in the IL-28B Gene
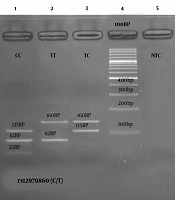
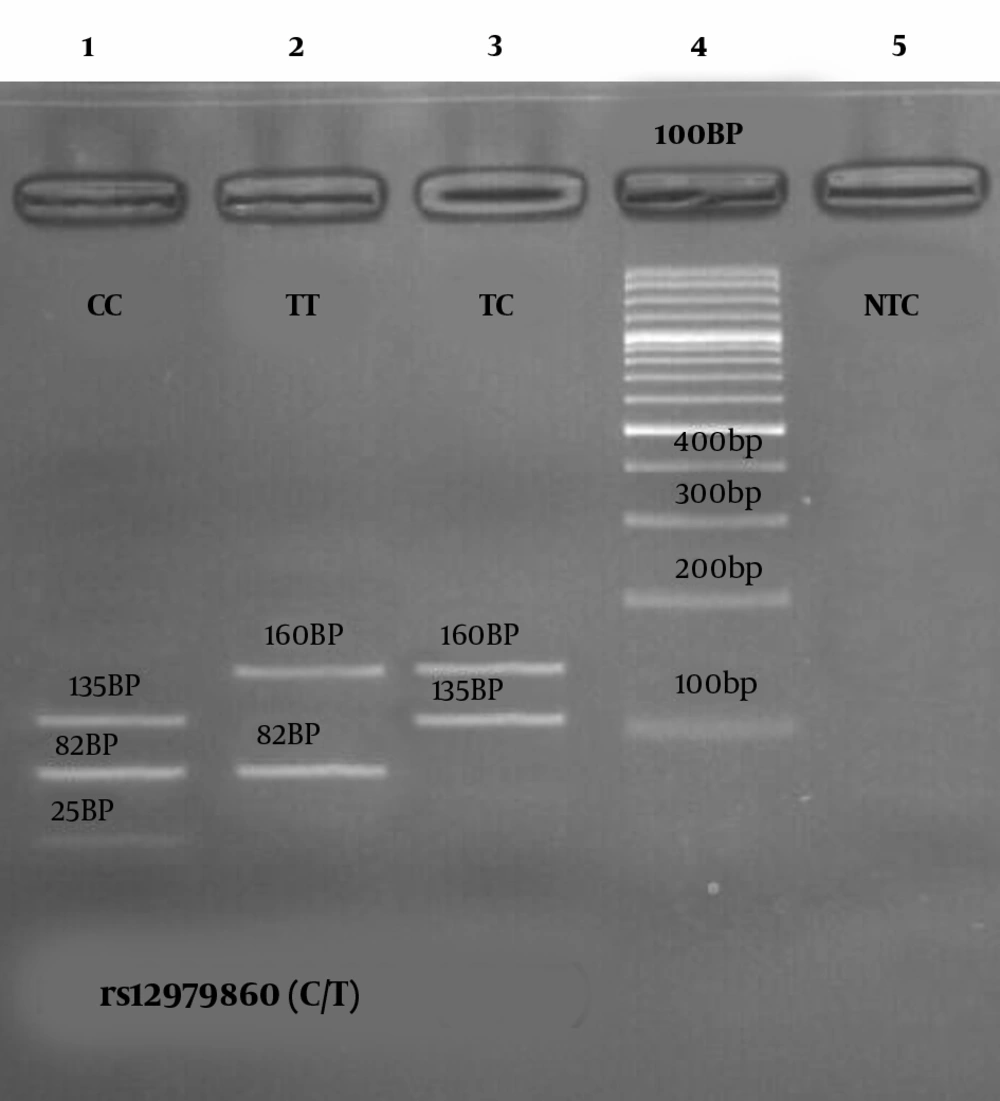
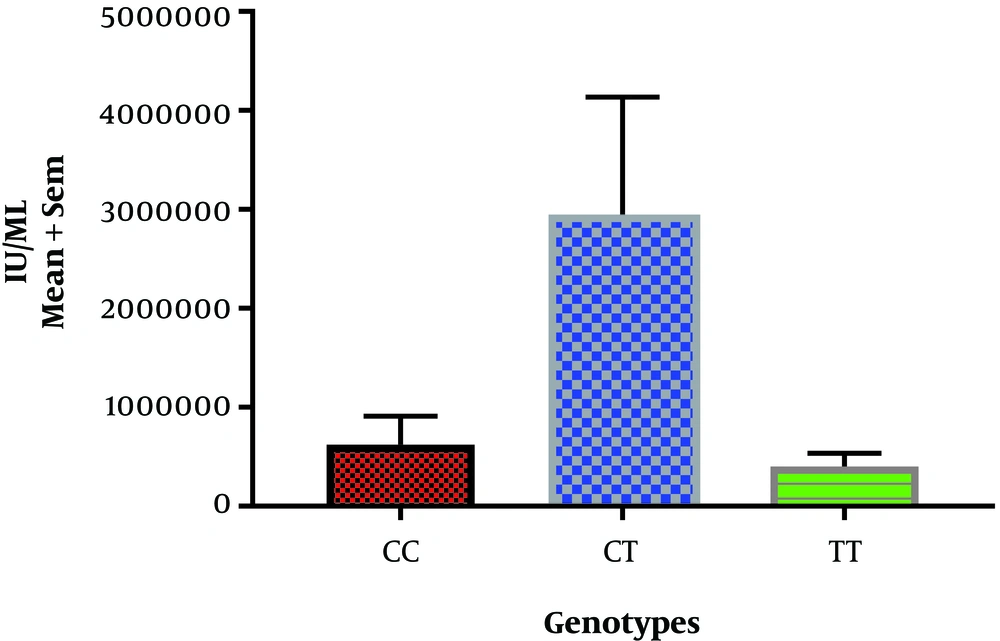
In this study, the genotype rs12979860 correlation with gender was statistically significant for the control group and HCV patients as well (P < 0.033). From HCV patients, 40 cases (33.3%) had Genotype 1 (CC), 52 cases had Genotype 2 (CT) (43.3%), and 28 cases had Genotype 3 (TT) (23.3%). In general, the risk of infection for CC genotypes was not statistically significant (P < 0.103). However, in the control group, genotype 1 was seen in 60 persons (50%), genotypes 2 in 39 persons (32.5%), and genotypes 3 in 21 persons (17.5%) (Table 3). After performing the PCR, a 241 bp fragment was obtained (Figure 1 of well 1) from the rs12979860 polymorphism-contained location of the samples which displayed three genotypes after digestion as it follows: The CC genotype showed three parts of 135bp, 82bp, and 25bp (Figure 1, well 1). The TT genotype showed two parts of 82bp and 160bp (Figure 1, well 2). The TC genotype also showed two pieces of 135bp and 160bp (Figure 1, well 3). The relationship between the viral load and the different genotypes is also shown in Figure 2.
| Genotypes | Control (N = 120) | HCV Patient (N = 120) | Total (N = 240) | P Value |
|---|---|---|---|---|
| Codominant | ||||
| CC | 60 (50.00) | 40 (33.30) | 100 (41.70) | 0.009 |
| CT | 39 (32.50) | 52 (43.30) | 91.00 (38.20) | 0.103 |
| TT | 21 (17.50) | 28 (23.30) | 49.00 (20.40) | 0.22 |
| Dominant | 0.009 | |||
| CC | 59 (50) | 40 (33.3) | 99 (41.6) | |
| TT + CT | 59 (50) | 80 (66.6) | 139 (58.4) | |
| Recessive | 0.221 | |||
| TT | 20 (16.9) | 28 (23.3) | 48 (20.2) | |
| CT + CC | 98 (83.1) | 92 (76.6) | 190 (79.8) |
5. Discussion
Hepatitis C is one of the main causes of liver disease (22) with a small global prevalence. However, only a small number of infected people automatically clear themselves of the virus, and about 70 to 80 percent of patients become chronic carriers, which may lead to cirrhosis and Hepatocellular carcinogenesis. Also, in hepatitis C, the patient's response to the treatment is different, and its treatment is difficult to tolerate (23). In the treatment of hepatitis C, various factors can be helpful, including the type of treatment, drugs, and effective factors in treating antiviral containing the viral and host factors (24). Moreover, the role of the IL 28B gene in SVR of hepatitis C disease is notable (25).
IL 28B polymorphism is more potent; it can predict the response to treatment better than other pre-emerging predictor factors, such as racial background, initial viral load, liver fibrosis, fasting glucose, and BMI, in Hepatitis C patients (26). Previous findings have also shown that rs12979860 SNP genotype in IL 28B gene plays a determining role in response to treatment (27). Also, only 20 - 30% of the contaminated cases show clinical symptoms (28, 29). Whereas, 70 - 80% of infected cases develop chronic infections, and 30% of those with chronic infection develop chronic liver disease, including progressive fibrosis, resulting in cirrhosis and hepatocellular carcinoma (12).
Understanding the importance of this genotype and its role in the SVR of hepatitis C patients can help determine the extent to which it can be cured in the population. The prevalence of this disease varies in different regions of Iran, with different results reported in the previous study. In the Isfahan study, TT, CT, and CC genotypes were 17%, 41%, and 42%, respectively (30). On the other hand, the study in Yazd showed that the highest IL-28 gene level in HCV patients was related to CT genotype (55.4%) (20). Also, the prevalence of CC, CT, and TT genotypes in another study in Mazandaran was 41.4%, 41.6%, and 17.3%, respectively (21). Despite the difference in the frequency of genotypes examined by IL 28B SNP rs12979860, the optimal C allele is more prevalent in different populations. The results of our study showed that the CC genotype had a lower proportion in HCV patients of both genders than in the control group.
On the other hand, the optimal C allele in healthy subjects was 66.6% and less frequent in patients (55%). According to the previous studies, CC genotype can increase the resistance to HCV treatment in patients (31, 32), which is confirmed by the result of the present study (P = 0.009). The findings from different populations indicate the specific role of this genotype in HCV patients. One of these studies was performed in a population of HCV patients in Turkey, where it was found that the CC genotype in the control group was higher than in the patient group, and the difference between CC and TT genotypes’ frequency was significant (P = 0.016) (33).
Therefore, studies in different populations address the importance of the CC allele in a desirable and effective response to treatment in HCV patients (20, 21, 30, 34). Based on the epidemiologic studies, the prevalence of this allele is greater in the population of East Asian people, so they are probably more resistant to HCV. On the other hand, after East Asia, it has been mostly observed in South Asia, Europe, and Africa, respectively (12). Although individuals with CC genotype are more likely to be treated with PEG-IFN and RBV, race can affect the treatment results, and the rate of SVR in individuals with a CC genotype varies by race (35), though other factors that could affect the amount of SVR are not explicitly explained.
5.1. Conclusions
Given the importance of the desirable CC genotype in healthy individuals and patients, it can be suggested that detecting the rs12979860 polymorphism genotype of IL 28 gene can help determine the treatment process for HCV patients. However, a multi-dimensional and accurate evaluation is also required to design and conduct more extensive studies in larger scales through other regions of Iran to confer a more comprehensive report.