1. Background
Zika Virus (ZIKV) infection has recently caused a significant concern worldwide. This virus was first isolated from the serum of rhesus monkeys with fever in the Zika forest in April 1947 (1). After more than half a century, ZIKV was first detected in areas outside Africa and Asia (2). Recently, its outbreak in Asia, Africa, and America in 2018 caused many newborns with microcephaly symptoms, which raised much attention. Previous studies showed that the symptoms of ZIKV infection were actually asymptomatic and did not cause huge damage, without wide concerns. Now, there is growing evidence that ZIKV can cause some severe diseases, including fetal brain development disorders and Guillain-Barré syndrome (3). New evidence suggests that ZIKV infection may have a detrimental effect on the heart of some patients. In February 2016, the ZIKV infection was recognized as a public health emergency of international concern by the World Health Organization (WHO) (4).
However, ZIKV infection is still erupting in many areas such as Southeast Asia and Brazil. The cases of natural ZIKV infections have been reported by 46 countries, accounting for over 1.5 million people infected with this virus. The virus can be transmitted from humans to humans by Aedes species mosquitoes, but it can also infect humans through blood, mother-to-child transmission, and sexual contact (5-7). The ZIKV is a single-stranded, positive-sense RNA virus, closely similar to the Dengue Virus (DENV), Japanese Encephalitis Virus (JEV), and West Nile Virus (WNV) (8-10). The virus has a diameter of 40-70 nm that consists of three structural proteins and seven nonstructural proteins (11, 12). At present, ZIKV strains have evolved into the African and Asian lineages (13, 14). Several studies showed that the amino acid mutation of nonstructural protein 1 (NS1) on ZIKV Asian strain (NS1 A188V) promoted the secretion of the virus NS1 protein and enhanced the viral infectivity of A. aegypti mosquitoes, facilitating the ZIKV transmission and leading to recent large-scale epidemics (15, 16).
Now, infections with ZIKV are life-threatening. There is the possibility of spreading to other regions and potential epidemics in the future. An accurate biomarker is very important for early diagnosis, effective control of epidemics, and timely treatment. The NS1 protein, as a multifunctional pathogenicity factor, is the most enigmatic protein of flaviviruses (17). It is critical for RNA replication and immune evasion in flaviviruses (18). In previous studies of DENV, the NS1 protein was secreted from infected cells in the form of hexamers (19). The NS1 proteins with high immunogenicity are present in sera infected with DENV (20). Many studies reported that the NS1 protein was used as a marker to diagnose DENV infections. Besides, it is the important components of experimental vaccines (21).
Although ZIKV is similar to other flaviviruses, in terms of protein folding and domain alignment (10, 22), the virus has some unique structural features, such as the most variable outer surface of the wing domain (23). To learn more about the ZIKV-NS1 protein, the recombinant ZIKV-NS1 protein was expressed and used to prepare polyclonal antibody (pAb). In this study, the native NS1 protein secreted in a soluble form in supernatants of host cells infected with ZIKV was detected by our prepared antibodies. As the infection time increases, the amount of NS1 protein in the supernatant of host cells increases, too. Accordingly, we confirmed that the recombinant NS1 protein had good immunogenicity and the prepared antibody specifically reacted with the native NS1 protein produced in host cells infected with ZIKV. The prepared NS1 protein and antibody can play important roles in the detection of ZIKV. Besides, the recombinant NS1 protein and its antibody prepared in our study can contribute to developing NS1-based vaccines and NS1-targeting antibody to prevent flavivirus disease progression and reduce the harm to humans.
2. Objectives
This study aimed to express and purify the recombinant ZIKV-NS1 protein and prepare an antibody. Besides, we assessed the prepared antibody for efficient reaction with the native ZIKV-NS1 protein and its role in the accurate detection of ZIKV infections.
3. Methods
3.1. Virus, Cells, and Antibodies
ZIKV strain SZ-WIV01 (GenBank, NCBI accession KU963796) was originally isolated from a Chinese male patient and propagated in A. albopictus C6/36 cells. Escherichia coli, JM109, Rosetta (DE3), BHK-21, Vero, A. albopictus C6/36 cells were stored in our lab. Freund’s Incomplete Adjuvant (FIA) and Freund’s Complete Adjuvant (FCA) were purchased from Sigma (St. Louis, MO, USA). The goat anti-mouse IgG (H + L) HRP and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G were purchased from Abcam (Cambridge, UK). Ni-IDA beads and a MAg25K/IDA Ni kit were purchased from Enriching Biotechnology (Su Zhou, China). Protein A-sepharose was purchased from GE Healthcare (Chicago, IL, USA).
3.2. Construction of ZIKV-NS1 Prokaryotic Expression Vector
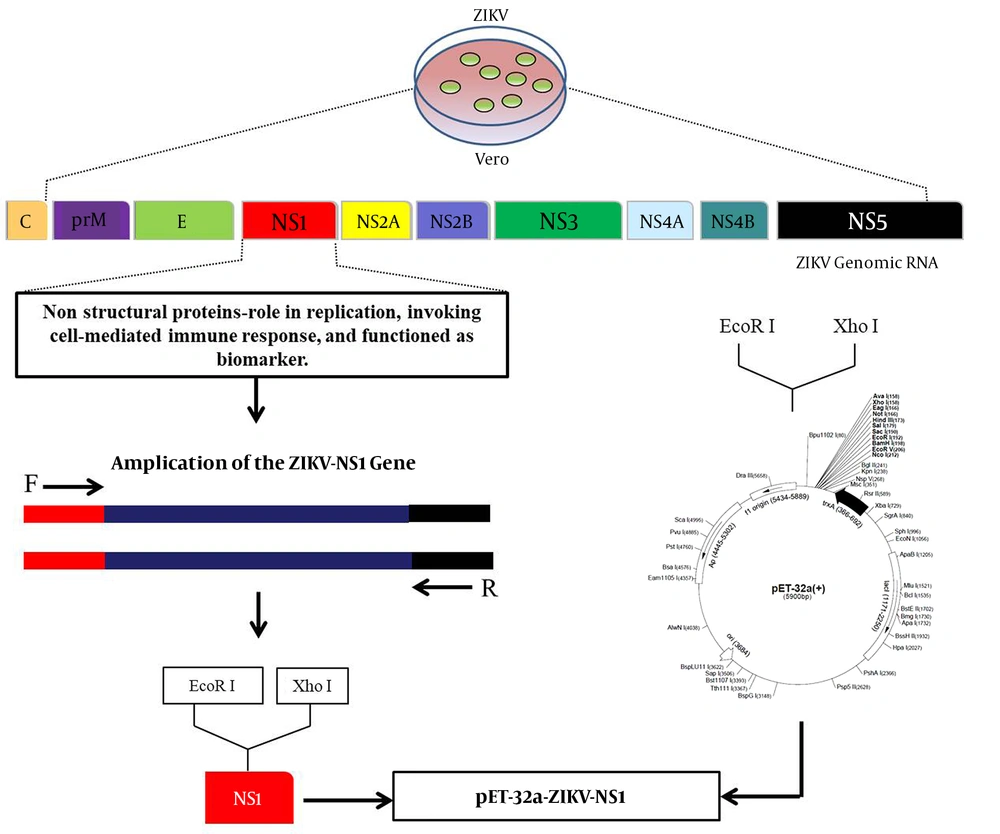
The ZIKV-NS1 prokaryotic expression vector was constructed as shown in Figure 1. Briefly, the supernatant of ZIKV-infected Vero cells was collected and the total RNA of ZIKV was extracted using a Pure Link® RNA Mini Kit (Life Technologies, USA). The specific primers F 5’ GGAATTCGATGTTGGGTGTTCAGT 3’ and R 5’ CCGCTCGAGTTACGCTGTCACCACAGACCT 3’ were performed to amplify the ZIKV-NS1 gene. The positive PCR products were gel-purified using a Gel Extraction Mini Kit (Tiangen, China). The gel-purified ZIKV-NS1 gene was digested with the EcoR I and Xho I enzymes and ligated into the pET-32a (+) vector digested with the same enzymes to generate pET-32a-NS1.
3.3. Expression and Purification of ZIKV-NS1 Recombinant Protein
The recombinant plasmid of pET-32a-NS1 was transformed into E. coli Rosetta and coated on an LB Ampicillin (AMP) solid plate. After being confirmed by bacterial PCR and DNA sequencing, the positive clone was cultured in an LB liquid medium containing AMP (100 μg.mL-1) for 6 h at 37ºC with shaking at 180 rpm. Then, the ZIKV-NS1 protein was induced with isopropyl-β-D-thiogalactoside (IPTG) for 12 h at 22ºC with shaking at 120 rpm for a final concentration of 0.1 mM. Thereafter, the precipitate was collected and suspended in 50 mL of binding buffer (20 mM sodium phosphate, 500 mM NaCl, 1 mM PMSF, and pH 7.4). The cells were disrupted on ice using a sonicator. Meanwhile, the Ni-IDA beads were mixed evenly, then washed with deionized water and binding buffer. Next, the crude proteins were mixed with the as-prepared Ni-IDA beads for 30 min at room temperature. Thirty minutes later, the supernatant was removed by magnetic separation and the beads were washed three times with wash buffer (20 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole, and pH 7.4). After washing, the NS1 proteins were collected after adding 3 mL of elution buffer (20 mM sodium phosphate, 500 mM NaCl, 400 mM imidazole, and pH 7.4) with shaking for 10 min at 100 rpm. Lastly, the purified proteins were analyzed by SDS-PAGE.
3.4. Preparation of pAb Against Recombinant ZIKV-NS1 Protein
The pAb against the ZIKV-NS1 protein was performed according to Zhang et al. (24). The prepared NS1 protein was used to immunize BALB/c mice, with 30 µg of NS1 protein mixed with FCA for the first immunization and 50 µg of NS1 protein mixed with FIA for the second and third immunizations. Seven days after the third immunization, the boosted immunization was performed by injecting the ZIKV-NS1 protein through the tail vein and the polyclonal antisera against ZIKV-NS1 were prepared successfully. Then, the Protein A Sepharose (GE Healthcare) was used to purify the Immunoglobulin G (IgG) of pAb (25).
3.5. Titer of pAb
The titer of the pAb was determined by Enzyme-linked Immunosorbent Assay (ELISA) after the anti-serum was purified (26). Briefly, 1 μg of NS1 protein was coated on each well and maintained overnight at 4ºC. Then, the plate was blocked for 1 h at 37ºC with the blocking buffer (3% BSA in PBS). Next, the pAb was diluted from 1:200 to 1:204,800; then, 100 µL of each dilution was added to each well and maintained for 2 h at 37ºC. Thereafter, the goat anti-mouse IgG (H+L) HRP (1:5000) was added and incubated for 1 h at 37ºC. After each step, the wells were washed three times with phosphate-buffered saline containing 0.05% Tween buffer (PBS-T). After washing, the TMB soluble substrate (TIANGEN, China) was added to detect the immune response. Finally, the immune reaction was terminated by the stop buffer (2 M H2SO4) and the value of OD 450 was measured using a microplate reader (Bio-Rad, USA).
3.6. Western Blot
The activity of pAb was evaluated by Western blot. The native NS1 protein was prepared in ZIKV-infected BHK-21 cells while uninfected BHK-21 cells were used as a control. All proteins were collected and transferred to a Nitrocellulose (NC) membrane. Then, the membrane was blocked for 2 h at 37ºC with blocking buffer. Next, the pAb (1:1000) was added to react with the native NS1 protein on the membrane for 2 h at 37ºC. Two hours later, the membrane was incubated with HRP-conjugated secondary antibodies (1:5000) for 1 h at 37ºC. After each incubation step, the membrane was washed with PBS-T as described above. Finally, the immune response of pAb was detected using a Western blot kit (BioBest, Anhui, China).
3.7. Indirect Immunofluorescent Assay
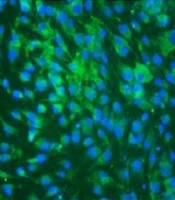
For Indirect Immunofluorescent Assay (IFA), ZIKV-infected BHK-21 cells and uninfected BHK-21 cells were fixed with 4% Paraformaldehyde (PFA) for 30 min at 4ºC. Next, the cells were blocked with blocking buffer and incubated with pAb (1:1000). Thereafter, the FITC-conjugated goat anti-mouse immunoglobulin G (1:1000) was added and incubated for 1 h at 37ºC. After each incubation step, each well was washed with PBS-T as mentioned above. Finally, each well was added with 200 µL PBS for fluorescence detection.
3.8. Prediction Methods
The iLoc-Virus (http://jci-bioinfo.cn/iLoc-Virus) and Virus-mPLoc (http: //www.csbio.sjtu.edu.cn/bioinf/virus-multi/), as the two commonly used protein subcellular localization programs, were used for the prediction of ZIKV-NS1 protein. These two programs can predict eukaryotic proteins via web servers and accept a large number of sequences. Besides, both programs have the ability to predicate subcellular locations, including Host Nucleus (HNuc), Viral Capsid (VC), Host Peroxisome (HPer), Secreted (Sec), Host Cytoplasm (HCyt), Host Cytoskeleton (HCyk), Host Endoplasmic Reticulum (HER), Host Golgi-apparatus (HGA), Cell Membrane (HCM Host), Host Mitochondrion (HMit), Host Lysosome (HLys), Extracell (Ext), and Elsewhere (Ew).
3.9. Application of ELISA and Western Blot for Detection of NS1 Proteins Secreted in Supernatant of ZIKV-Infected Cells
Briefly, ZIKV was used to infect Vero, BHK-21 cells (MOI = 1). The infected cells were cultured in the DMEM medium, containing 1% (v/v) FBS. The supernatants of ZIKV-infected Vero and BHK-21 cells were collected and concentrated from various infection periods (12 h, 24 h, 36 h, 48 h, 60 h, and 72 h). Then, the supernatants of ZIKV-infected Vero, BHK-21 cells from different infection periods were used for ELISA and Western blot assay as described above.
4. Results
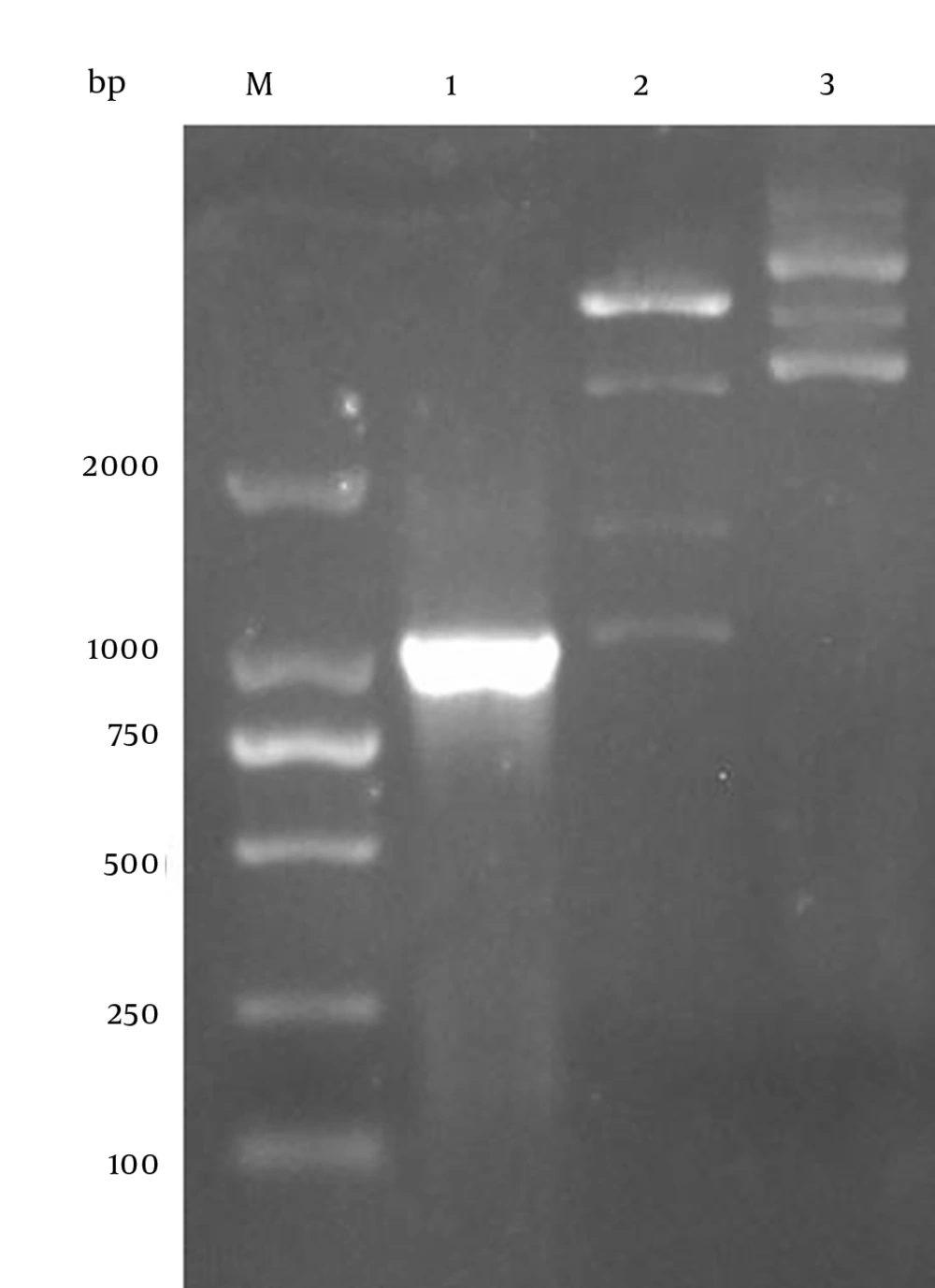
4.1. Generation of ZIKV-NS1 Protein Prokaryotic Expression Plasmid
The RNA of the SZ-WIV01 strain was extracted and amplified by PCR (Figure 2, lane 1). Next, the PCR products were gel-purified and inserted into fraction pET-32a (+) digested with EcoR I and Xho I. Constructed recombinant plasmid pET-32a-NS1 was confirmed by bacterial PCR, restriction enzyme digestion, and DNA sequencing (Figure 2, lanes 1 and 2). Besides, the pET-32a vector was digested by the same enzymes as a control (Figure 2, lane 3).
4.2. Expression and Purification of Recombinant ZIKV-NS1 Protein
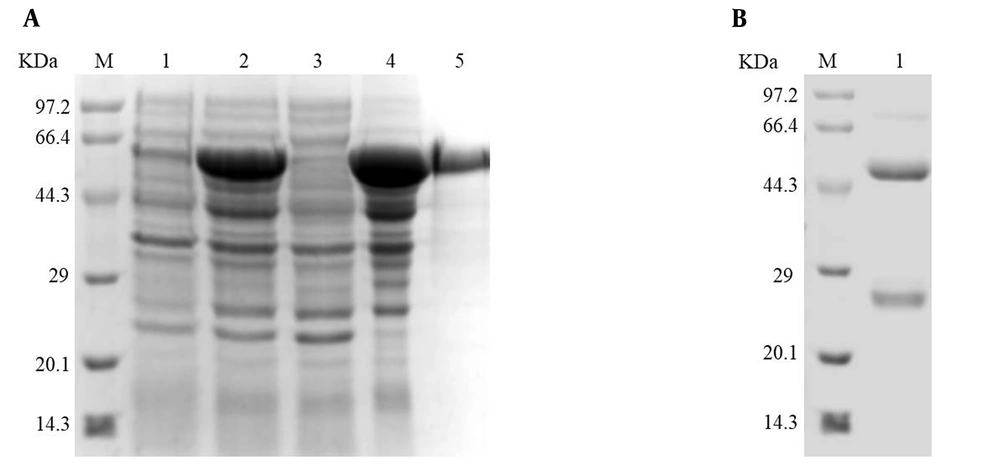
The recombinant ZIKV-NS1 protein was expressed at a relatively high level through BL21 (DE3) cells when induced with IPTG (Figure 3A, lane 2). The solubility of the recombinant ZIKV-NS1 protein was confirmed by SDS-PAGE (Figure 3A, lanes 3 and 4). The SDS-PAGE revealed that the ZIKV-NS1 protein was mainly expressed in the cell debris pellets in the form of inclusion bodies (Figure 3A, lane 4). Then, the inclusion bodies were denatured by 8 M urea (8 M urea, 50 mM Tris-HCl, 100 mM NaCl, and pH 7.9) and the recombinant NS1 protein was purified by Ni-IDA beads (Figure 3A, lane 5). Finally, the recombinant ZIKV-NS1 protein was refolded in PBS buffer.
Expressed and purified ZIKV-NS1 protein and pAb. (A) Lane 1: Escherichia coli lysate before induction; lane 2: E. coli lysate after induction; lane 3: soluble proteins after centrifugation; lane 4: insoluble proteins after centrifugation; lane 5: recombinant ZIKV-NS1 protein after purification. (B) Purified pAb using Protein A-sepharose
4.3. Evaluation of Antisera
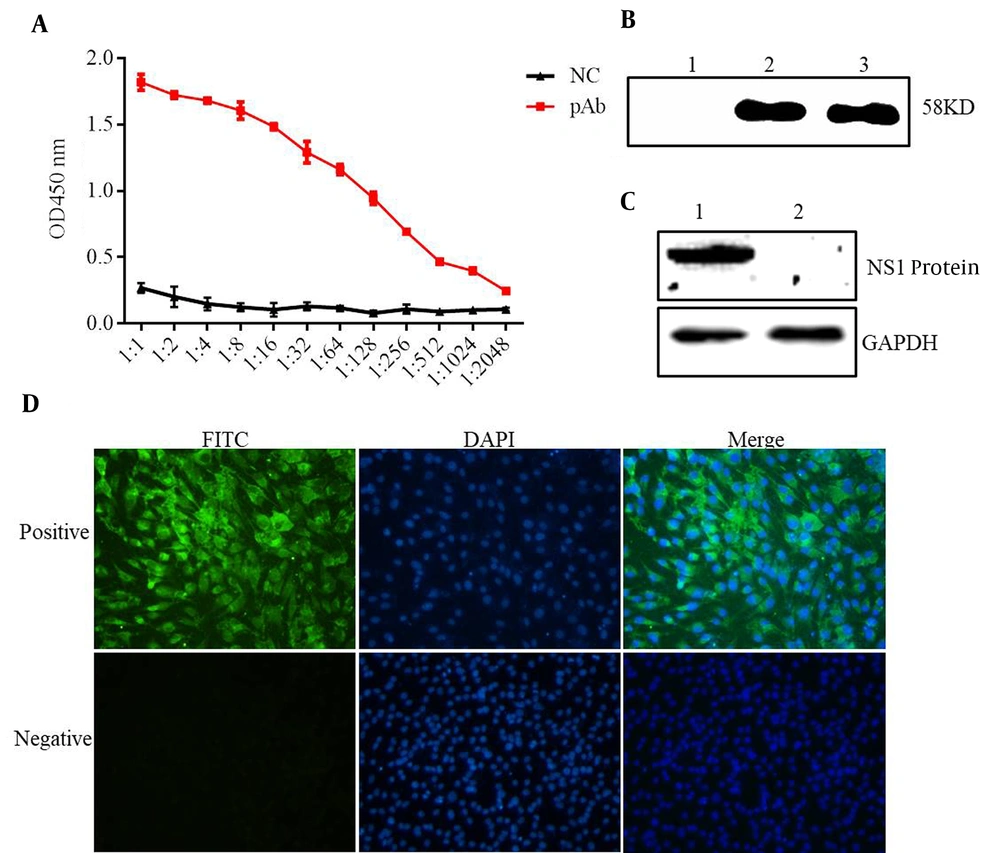
The purified NS1 protein was used as an antigen to immune mice. After three times of normal immunization and booster immunization, the antisera were prepared and purified using Protein A-sepharose. The purified immunoglobulin was confirmed by SDS-PAGE (Figure 3B, lane 1). The titer of antisera was determined by ELISA (Figure 4A). The reactivity and specificity of the antisera were evaluated by Western blot and IFA. Figure 4B and C showed that the pAb could recognize both recombinant ZIKV-NS1 protein and native NS1 protein in ZIKV-infected cells specifically. Figure 4D, consistent with Figure 4B and C, showed that the anti-ZIKV-NS1 serum could react with the native NS1 protein in ZIKV-infected Vero and BHK-21 cells while the negative control was not recognized.
Evaluation of pAb against recombinant ZIKV-NS1 protein. (A) ELISA of the titer of pAb. (B) Western blot assay of pAb; lane 1: Escherichia coli without IPTG; lane 2: induced ZIKV-NS1 protein; lane 3: purified recombinant ZIKV-NS1 protein. (C) Native ZIKV-NS1 protein in ZIKV-infected BHK-21 cells detected for Western bot analysis and the mock-infected BHK cells used as a control. (D) Indirect immunofluorescence assay of pAb
4.4. Prediction Results for ZIKV-NS1 Protein
According to the website instruction, the nucleic acid sequence of ZIKV-NS1 protein was added in the iLoc-Virus and Virus-mPLoc proteins for the prediction of subcellular localization. The results of the two subcellular localization programs showed that the NS1 protein could be secreted extracellularly, as shown in Table 1.
| Programs | HCyt | HER | HGA | HCM | HMit | Hnuc | HPer | Sec/Ext | VC | HLys | EW |
|---|---|---|---|---|---|---|---|---|---|---|---|
| iLoc-Virus | √ | √ | √ | √ | √ | √ | |||||
| Virus-mPLoc | √ | √ | □ | √ | □ | √ | □ | √ | √ | □ | □ |
Predictable Subcellular Locations of ZIKV-NS1 Protein Using Virus-PLoc and Virus-mPLo
4.5. Detection of Native NS1 Proteins by ELISA and Western Blot
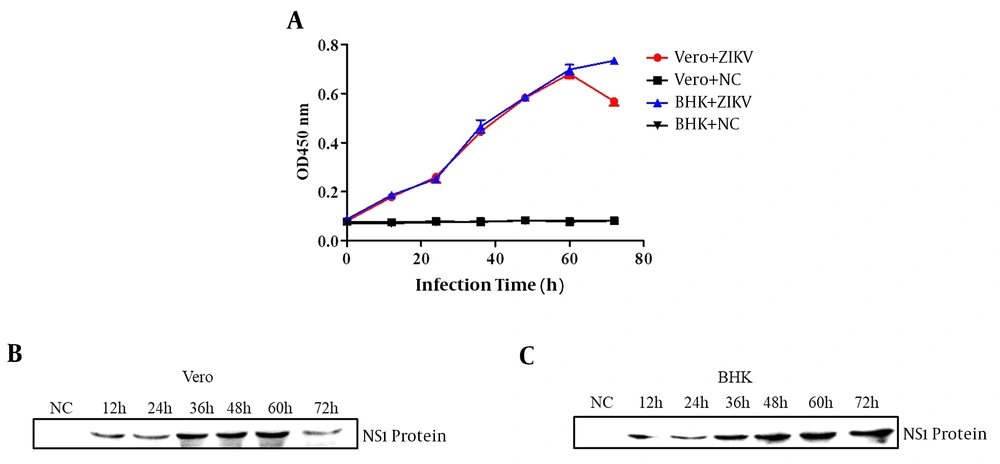
To verify that the antibody against the recombinant ZIKV-NS1 protein could react with the native NS1 protein secreted by ZIKV-infected host cells, the Vero and BHK-21 cells were infected with ZIKV for various periods. Next, the supernatants of the infected cells were collected at various times and used for ELISA and Western blot assay. The results of ELISA proved that the amount of native NS1 proteins secreted in the supernatant of infected cells gradually increased and it was detectable at various infection periods from 12 h to 72 h (Figure 5A). Besides, the Western blot confirmed that the native ZIKV-NS1 proteins were secreted in the supernatant. As shown in Figure 5B and C, as the infection duration increased from 12 h to 72 h, the target band was observed more clearly by the Western blot kit (BioBest, Anhui, China).
Soluble NS1 proteins detected by ELISA and Western blot. (A) The ELISA of native NS1 proteins secreted in the supernatant of ZIKV-infected Vero cells and BHK-21 cells. The supernatant of Vero cells (B) and BHK-21 cells (C) infected with ZIKV in various infection periods detected by Western blot
5. Discussion
The ZIKV, as a member of the Flaviviridae family, is one of the most threatening viruses. It was isolated in 1954 from patients in an outbreak of “jaundice” in West Africa. Only sporadic cases occurred in Africa and Asia and no large-scale outbreaks were observed (27). Recently, a large-scale outbreak of ZIKV in the northeast of Brazil caused a myriad of neurological disorders, leading to the number of babies born with microcephaly (28). The ZIKV infection poses a serious pandemic threat to over 46 countries and more than 1.5 million people have been confirmed to be infected with ZIKV worldwide. The early diagnosis of ZIKV infection would be more effective in controlling epidemics and timely treatment (29).
Currently, no effective drugs are available for ZIKV infection and a vaccine is still under development. Since the ZIKV infection is actually asymptomatic, it is difficult to be diagnosed in early stages based on clinical manifestations. Thus, it is rare to diagnose patients more than 1 to 2 weeks of post-symptom onset (30, 31). The early diagnosis of ZIKV infection would be more effective in controlling epidemics and timely treatment. Many detection methods have been reported to diagnose ZIKV infection during recent outbreaks, including the fluorescence quantitative Polymerase Chain Reaction (RT-qPCR) (32), serological methods (e.g., IgG and IgM antibodies) (33), and loop-mediated isothermal amplification (LAMP) assay (34). However, the short duration of the viremic phase leads to a narrow diagnostic window for the detection of viral components. Therefore, the detection methods based on viral RNAs such as RT-qPCR and LAMP are not accurate. Furthermore, IgM/IgG is usually generated about seven days after symptom onset and it is variable in different infected patients. Thus, it is not suitable for the early diagnosis of ZIKV.
In this study, we focused on the biochemical characterization of an antibody against the recombinant ZIKV-NS1 protein. Prokaryotic expression vector pET-32a-NS1 was constructed and subsequently, the recombinant ZIKV-NS1 proteins were purified successfully. To evaluate that the antibody against the recombinant ZIKV-NS1 protein could react with the native NS1 protein, the recombinant ZIKV-NS1 was used to immunize mice. Both Western blot and IFA results showed that the antibody against recombinant ZIKV-NS1 could recognize the native NS1 protein in ZIKV-infected cells. Moreover, we used two commonly used protein subcellular localization programs to predicate the possibility of subcellular localization of the ZIKV-NS1 protein. Both methods revealed that the NS1 protein was secreted extracellularly. The NS1 protein also participated in viral replication. In this study, the different criteria were used to evaluate the usefulness of the recombinant NS1 antigen and its antibody. Based on the results of ELISA and Western blot, we found that the soluble NS1 protein was detected by our prepared antibody in various infection periods. Besides, our results also demonstrated that the NS1 protein could act as an ideal biomarker and could recognize the initial stage of ZIKV infection to monitor the progress of infection (35).
5.1. Conclusions
In conclusion, we found that the ZIKV-NS1 protein could be released in the supernatant of ZIKV-infected Vero, BHK cells in a soluble form. More importantly, the recombinant NS1 protein from a prokaryotic expression vector had good immunogenicity and the prepared antibody could efficiently recognize the native NS1 protein. This phenomenon was confirmed by ELISA, Western blot, and IFA. Our results revealed that the secreted native NS1 protein was detected by our antibody in various infection periods. Overall, we demonstrated that our prepared antibody is a reliable biological tool that enables the immunological detection of secreted NS1 from host cells infected with ZIKV. Our findings provide the basis for the development of rapid diagnostic kits such as sandwich ELISA and sandwich lateral flow strip, which can facilitate the monitoring of ZIKV in a variety of clinical samples and guide for clinical medication and timely treatment of patients effectively. Besides, the recombinant NS1 protein and its antibody would make contributions to developing NS1-based vaccines and NS1-targeting antibodies to inhibit NS1 activity and prevent flavivirus disease progression.