1. Background
Human metapneumovirus (HMPV), human parainfluenza viruses (HPIV-1, HPIV-2, HPIV-3, and HPIV-4), respiratory syncytial virus type A (RSV-A, RSV-B), influenza A and B viruses and HCoVs (OC43/HKU1, NL63, 229E) are common viruses to cause acute respiratory infection (ARI) worldwide (1). Human metapneumoviruses (HMPVs) are the members of the genus Metapneumovirus and family Pneumoviridae (2). Human metapneumoviruses are causative agents of ARI worldwide (3, 4). Human metapneumovirus is classified into two distinct groups A and B. Based on nucleotide sequence variations, each group is further divided into two subgroups (A1 and A2 in group A and B1 and B2 in group B). In addition, subgroup A2 is further classified into two distinct clades A2a and A2b (5).
Human respiratory syncytial virus (HRSV) types A and B belong to genus Orthopneumovirus and family Pneumoviridae (2). The human respiratory syncytial virus is responsible for bronchiolitis and pneumonia in infants and young children (6, 7). Human respiratory syncytial virus group A is subdivided into 11 genotypes such as GA1-GA7, ON1, SAA1, NA1, and NA and HRSV group B is categorized into genotypes GB1-GB4, SAB1-SAB3, SAB4, URU1, URU2, BA1-BA12, and THB (8, 9). Human parainfluenza virus (HPIV) 1-4 are belong to family Paramyxoviridae HPIV-1 and HPIV-3 belong to genus Respirovirus, and HPIV-2 and HPIV-4 to genus Rubulavirus (10). Human parainfluenza viruses are considered common causes of acute upper and lower respiratory infections among children and adult populations worldwide (11). Parainfluenza viruses (HPIVs) are associated with clinical illnesses such as otitis media, pharyngitis, conjunctivitis, croup, tracheobronchitis, and pneumonia based on genetic and antigenic variations (polymerase, matrix, and nucleoprotein). Human parainfluenza viruses are classified into four types HPIV-1 to HPIV-4, among which HPIV-4 is regarded as less important (11).
There are four types of influenza viruses A, B, C, and D. Human influenza A (H1N1, H3N2) and B viruses can cause seasonal epidemics of diseases including ARI among all age-groups in different regions of the world (12, 13). Four types of HCoVs (HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1) have been associated with a wide range of respiratory illnesses, including the common cold, pneumonia, and bronchiolitis (14). Concerning clinical manifestations of ARI, it is difficult to distinguish between RSV, HMPV, influenza virus (A, B), HPIVs (1-4) and HCoVs (OC43/HKU1, NL63, 229E) infections. Therefore, the primary diagnosis is important to ease the early management and control of ARIs (15).
To date, the standard detection of viral ARI has been achieved through the application of molecular methods. Among them, the real-time PCR technique provides a productive solution for early diagnosis of specific respiratory DNA or RNA viruses (16). To compare real-time PCR with traditional virus culture and immunofluorescence detection methods, real-time PCR has advantages of highly sensitive, rapid, and simultaneous detection of multiple pathogens. Multiplex real-time PCR allows detecting multiple pathogens simultaneously in a single reaction as a cost-effective method (17).
2. Objectives
This study was conducted to evaluate the detection of HMPV, RSV-A, RSV-B, HPIV-1, HPIV-2, HPIV-3, HPIV-4, influenza A and B viruses, and HCoVs (OC43/HKU1, NL63, 229E) in hospitalized patients using multiplex real-time PCR.
3. Methods
3.1. Study Design and Sample Collection
The samples for this cross-sectional study included throat swabs received from hospitals of Ahvaz city, Iran, from September 2015 to December 2016. The target population comprised adults above 40 years of age who referred to the hospitals with clinical ARI. Most patients had symptoms, including congestion, runny nose, coughs, sore throat, body aches, fatigue, fevers over 39°C, chills, difficulty in breathing, dizziness, loss of consciousness, bronchitis, pneumonia, and bronchiolitis. Throat swabs were obtained from patients, put in viral transport medium with ice packs, and transferred to a Virology Department. They were kept at -80°C before RNA extraction.
3.2. RNA Extraction and cDNA Synthesis
Viral RNA was extracted from 112 throat swabs using a high-pure nucleic acid extraction kit (Roche Diagnostic, Manheim, Germany) following the manufacturer’s instructions. The cDNA was synthesized from the extracted RNA samples using a thermos-scientific cDNA synthesis kit following the manufacturer’s instructions.
3.3. Primers and Real-Time PCR
Based on TaqMan technology, real-time PCR was used to detect viruses associated with ARI. Table 1 shows primers and probes used for different viral respiratory infections including influenza A (IfA) H1N1 and H3N2 and influenza B (IfB) viruses, parainfluenza viruses (HPIV-1, HPIV-2, HPIV-3, and HPIV-4), respiratory syncytial viruses (RSVs) A and B, human coronavirus HCoVs (OC43/HKU1, NL63, 229E), and human metapneumoviruses A and B. The PCR reaction mixture contained 2X reaction buffer, 10 µM of each of forward and reverse primers, 2 µL of cDNA template, and D/W water up to 20 µL. All the samples including positive and negative controls were subjected to ABI one-step thermal cycling with the following conditions: initial denaturation at 95°C for 5 minutes, followed by 40 cycles of 95°C for 30 seconds and annealing at 60°C for 20 seconds.
| Virus (Gene Target) | Amplicon Size (bp) | Function (5/3 Probe Dye) | Oligonucleotide Sequence |
|---|---|---|---|
| RSV polymerase | 94 | Forward, L gene, 13907 - 13934 (MH447960.1) | AATACAGCCAAATCTAACCAACTTTACA |
| Reverse,14000 - 13980 | GCCAAGGAAGCATGCAATAAA | ||
| Probe A (FAM/MGB) | TGCTAYTGTGCACTAAAGa | ||
| Probe B (VIC/MGB) (MK534512.1) | CACTATTCCTTACTAAAGATGTC | ||
| MPV fusion (F) | A = 69 | Forward F, 3580 - 3602, (KJ627430.1) | GCCGTTAGCTTCAGTCAATTCAA |
| Reverse F, 3627 - 3648 | TCCAGCATTGTCTGAAAATTGC | ||
| Probe A (FAM/MGB) | CAACATTTAGAAACCTTCT | ||
| B = 74 | Forward, 3597 - 3619 (KF530178.1) | GCTGTCAGCTTCAGTCAATTCAA | |
| Reverse, 3648 - 3670 | GTTATCCCTGCATTGTCTGAAAACT | ||
| Probe B (VIC/MGB) | CGCACAACATTTAGGAATCTTCT | ||
| P1V1 (L protein) | 84 | Forward, L,12605 - 12634 (MH892404.1) | ACAGATGAARTTTTCAAGTGCTACTTTAGTa |
| Reverse, 12660 - 12688 | GCCTCTTTTAATGCCATATTATCATTAGA | ||
| Probe B (FAM/BHQ) 12638 - 12658 | ATGGTAATAAATCGACTCGCC | ||
| PIV2 (L protein) | 78 | Forward L, 10773 - 10802 (KY674972.1) | TGC ATGTTTTATAACTACTGATCT TGCTAA |
| Reverse, 10828 - 10850 | GTTCGAGCAAAATGGATTATGGT | ||
| Probe B (VIC/MGB) | ACTGTCTTCAATGGAGATAT | ||
| PIV3 (matrix) | 66 | Forward M, 4172 - 4190 | TGCTGTTCGATGCCAACAA |
| Reverse, 4210 - 4237 | ATTTTATGCTCCTATCTAGTGGAAGACA | ||
| Probe B (FAM/MGB) | TTGCTCTTGCTCCTCA | ||
| PIV4A (nucleoprotein) | 72 | Forward 4A, 365 - 389 | GRGCATTATTATCTCTGCTTTCCTTa |
| Reverse 4A, 415 - 435 (KF878965.2) | GGCTCTGGCAGCAATCATAAG | ||
| Probe A (VIC/MGB), 400 - 416 | CACATCAATGCAGAATC | ||
| Forward 4B, 416 - 436 | GGAGCGTTATTATCTCTGCTTTCTTT | ||
| Reverse 4Bm, 397 - 413 (EU627591.1) | GGCTCTGGCAGCAATCATAAG | ||
| Probe B | CACATCAATGCAGAATC | ||
| FluA (matrix) | 111/112 | H1N1, matrix, 123 - 148. (HE584759.1, H1N1) | TCTYATGGAATGGCTAAAGACAAGACa |
| Forward H3N2, matrix, 137 - 161 (CY202867.1, H3N2) | CTYATGGAATGGCTAAAGACAAGACa | ||
| Reverse A | SCGTCTACGCTGCAGTCCTCa | ||
| Probe H1N1 (VIC/MGB) | CTGGGCACGGTGAGC | ||
| Probe H3N2 (FAM) | ACTGGGCACGGTGAG | ||
| Flu B (matrix) | 76 | Forward B, matrix, 25 - 46, (MK676289.1) | CACAATTGCCTACCTGCTTTCA |
| Reverse B, & 3 - 100 | CCAACAGTGTAATTTTTCTGCTAGTTCT | ||
| Probe B (VIC/MGB) | CTTTGCCTTCTCCATCTT | ||
| C0V (polymerase) | OC43/HKU1 = 96 | Forward 1, 15068 - 15087, OC43 (MK303625.1) HKU1, MK167038.1 | TGGTGGCTGGGATGATATGT |
| Reverse 1, 15141 - 15163 | GGCATAGCACGATCACACTTAGG | ||
| NL63 = 100 | Forward 2, NL63, (MG428707.1 ) | TTTATGGTGGTTGGAATAATATGTTG | |
| Reverse 2 | GGCAAAGCTCTATCACATTTGG | ||
| Probe 1A (FAM/MGB) | ATAATCCCAACCCATRAGa | ||
| 229E = 99 | Forward 3, 13973 - 13992, (KF293666.1) | TGGCGGGTGGGATAATATGT | |
| Reverse 3, 14046 - 14071 | GAGGGCATAGCTCTATCACACTTAGG | ||
| Probe 2 (VIC/MGB) | ATAGTCCCATCCCATCAA |
Primers and Probes Used to Detect Viruses Associated with ARI
3.4. Statistical Analysis
The data were analyzed using SPSS version 21.0 (SPSS Inc., Chicago, IL, USA). The chi-square test was used for the analysis of categorical variables including gender, distribution of viral infections among genders and different age groups. P values of ≤ 0.05 were considered statistically significant.
4. Results
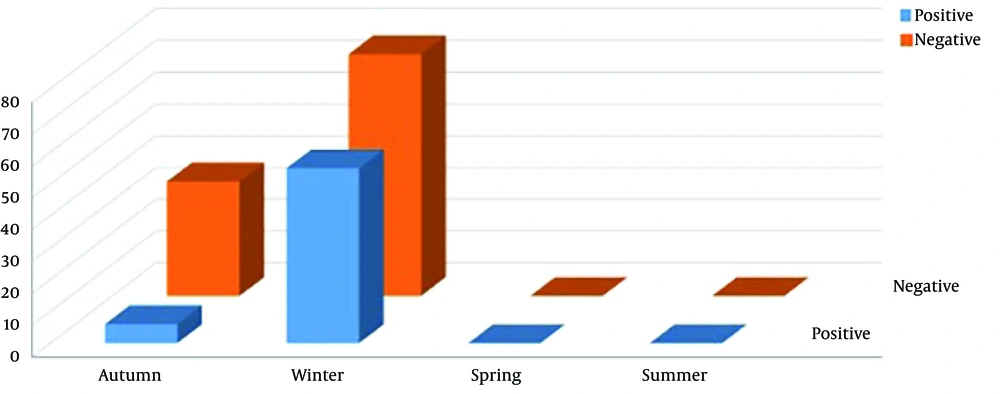
Within 15 months, 112 ARI patients were enrolled in the study. Among them, 55 (49.1%) were males and 57 (50.89%) females. Out of 112 samples, 61 (54.46%) were positive for viruses causing respiratory infections. Among them, there were 30 (26.78%) males and 31 (27.67%) females (P = 0.862). The frequency of viruses among ARI patients was as follows: Influenza A H3N2, n = 9 (8.03%), Influenza B, n = 1 (0.89%), RSVA, n = 28 (25%), MPVA, n = 18 (16.07%), HPIV1, n = 2 (1.78%), and PIV3, n = 3 (2.67%) (Table 2). Two (1.78%) specimens were simultaneously positive for two agents: RSVA/MPVA and RSVA/PIV3. The remaining 51 (45.53%) patients were negative for RSVB, MPVB, IfV H1N1, PIV2-4A-4B, and HCoVs (OC43/HKU1, NL63, 229E). Table 2 shows the distribution of viruses causing respiratory infection among different age groups. High frequency of viruses causing respiratory infections were observed in winter 55/76 (49.1%), while low detection of viruses were observed in autumn 6/36(5.35%) (Figure 1). No virus infection was detected in spring and summer (P = 0.000). Figure 1 illustrates the distribution of positive and negative viruses causing respiratory infection in different seasons.
| Category | Male | Female | P Value |
|---|---|---|---|
| Gender | 30 (26.78) | 31 (27.67) | 0.862 |
| RSV | 15 (13.39) | 13 (11.6) | 0.877 |
| Influenza A2 | 5 (4.46) | 4 (3.57) | |
| Influenza B | - | 1 (0.89) | |
| MPVA | 8 (7.14) | 10 (8.92) | |
| HPIV1 | 1 (0.89) | 1 (0.89) | |
| HPIV3 | 1 (0.89) | 2 (1.78) | |
| Total | 30/55 (25.89) | 31/57 (27.68) | |
| Age | 0.521 | ||
| 40 - 49 | 5 (4.46) | 6 (5.35) | |
| 50 - 59 | 9 (8.03) | 14 (12.5) | |
| 60 - 69 | 12 (10.71) | 9 (8.03) | |
| > 70 | 4 (3.57) | 2 (1.78) |
Distribution of Respiratory Virus Infections in Genders and Age Groupsa
5. Discussion
To date, viruses causing respiratory infections have been easily detectable in clinical laboratories by multiplex PCR tests. Thus, it will be helpful to acquire epidemiological data to ameliorate patient management. In the present study, 54.46% of patients were positive for viruses causing ARI. Among them, the most frequent viruses were RSV-A (n = 28; 25%) and HMPV-A (n = 17; 15.17%). Previous studies indicated that RSVA was the most frequent etiologic agent in patients with ARI (18-20). Arjeyni et al. reported a high frequency of RSVA (81%) and RSVB (19%) in adult patients with ARI in Iran (21). Birger et al. detected 85/168 (50.6%) human rhinovirus (HRV), 65 (38.7%) coronavirus (CoV), and 18 (10.2%) other viruses (human adenovirus, human metapneumovirus, influenza virus, and parainfluenza virus) in patients with ARI in the USA (22). In our survey, the positivity of other viruses causing respiratory infections was as follows: H3N2 n = 9 (8.03%), Influenza B n = 1, (0.89%), HPIV-1 n = 2 (1.78%), and HPIV-3 n = 3 (2.67%).
Han et al. in South Korea detected high frequency of Rhinovirus (n = 17; 37%), followed by parainfluenza virus (n = 14; 30.4%), respiratory syncytial virus (n = 10; 21.7%), coronavirus (n = 4; 8.7%), human metapneumovirus (n = 3; 6.5%), adenovirus (n = 2; 4.3%), Influenza A virus (n = 1; 2.2%) while no Influenza B virus and human bocavirus were detected in patients with ARI (23). Han et al.’s findings are not consistent with our results. In our survey, 51 (45.53%) ARI patients were negative for RSVA, RSVB, Influenza A2, Influenza B, HPIV1, HPIV3, and HCoVs (OC43/HKU1, NL63, 229E). In this survey, we did not investigate the detection of human adenoviruses rhinovirus/enterovirus (RV/EV), human bocavirus (HBoV), and human parechovirus (HpeV) in patients with ARI.
In Iran, the circulation of Adenovirus (AdV) has been reported in patients with ARI. Pourakbari et al. reported that the frequency of AdV was 3.4% in patients with ARI in Tehran (24). Tabasi et al. described that the rate of the Human Boca virus was 10.7% in children with ARI in Tehran (25). In our study, HCoVs (OC43/HKU1, NL63, 229E) were not detected in ARI patients. Mehdi et al. reported that 5.5% of ARI patients were positive for Coronavirus E229 in Tehran (26). In our study, two (1.78%) patients had two mixed viral infections, RSVA/MPVA and RSVA/PIV3. Many researchers have described the co-infection of viruses causing respiratory tract infections. Yoshida et al. in Japan detected the co-infection of human rhinovirus, human metapneumovirus, and parainfluenza virus-3 in patients with ARI (27). Petrarca et al. in Italy reported the co-infection of HRSV and HRV in patients with lower respiratory tract infection (28). The distribution of viruses causing ARI were 30 (26.78%) in males and 31 (27.67%) in females (P = 0.862).
The influenza vaccination is not mandatory in Iran although high-risk groups need to receive the vaccine. In this experiment, the outbreaks of respiratory virus infections were observed in autumn and winter, which are consistent with other reports (29, 30). No virus causing respiratory infection was detected in spring and summer seasons in this region of Iran. This might be due to that the temperature exceeds 40°C in the spring and remains constant above 48°C during summer. Ribavirin and oseltamivir antiviral therapy can reduce the progression of URI to LRI and mortality rate due to RSV and influenza virus infections (31). Currently, no specific antiviral therapy is available against human rhinoviruses, human coronaviruses, human metapneumovirus, and human bocavirus. Therefore, new treatment and preventive options are required. Sensitive molecular methods like multiplex real-time PCR in clinical practice can help the rapid diagnosis of viruses causing respiratory infection and reduce improper antibiotic therapy (32). Overall, several investigations have described that multiplex real-time PCR as a non-invasive method is faster and more sensitive than other standard methods such as serology and culture (33, 34).
5.1. Conclusions
In conclusion, in this study, RSVA (n = 28, 25%) and MPVA (n = 18, 16.07%) were the most common causes of RI in adults. Considering the low diagnostic cost of clinical presentation and laboratory findings in ARI, multiplex real-time PCR as a rapid diagnostic test can be considered efficient in reducing the length of patients’ stay in hospitals. The results indicated that 49 (43.75%) patients were negative for RSVA, RSVB, Influenza A2, Influenza B, HPIV2, HPIV4, HCoVs (OC43/HKU1, NL63, 229E); however, it requires further investigation to disclose the role of human adenoviruses rhinovirus/enterovirus (RV/EV), human bocavirus (HBoV), and human parechovirus (HpeV).