1. Background
Patients having bacterial infections were successfully treating with antibiotics in the past. However, currently, the fast emergence of resistant bacteria and the absence of new drugs have presented a significant threat to human health (1). Antimicrobial resistance (AMR) is the ability of bacteria attained over time to show resistance to antibiotics causing untreatable infections resulting in prolonged illness, increased mortality rate, and high expenditure (1, 2). Urinary tract infection (UTI) is among the most prevalent bacterial infections, affecting about 150 million people annually worldwide. The overuse/misuse of antibiotics for this type of frequently occurring infections has contributed to the persistence of resistant pathogens (3-5).
Bacteria have developed different mechanisms, one of which is extended-spectrum β-lactamase (ESBL) that hydrolyzes the beta-lactam ring of a known class of beta-lactam antibiotics. Extended-spectrum β-lactamase is found in almost all species of Enterobacteriaceae, but its ratio is slightly higher in Klebsiella pneumoniae (6). In such a complex situation of Multidrug Resistance (MDR), colistin is considered the last resort antibiotic to date. On the other hand, colistin is being widely used in veterinary medicine that has already enhanced resistance to this antibiotic in bacteria (7, 8). In various studies, colistin resistance genes were found to be located on the chromosome, but recently, a plasmid-mediated colistin resistance gene, mcr-1, has been identified (7).
The emergence of plasmid-mediated colistin resistance due to the mcr-1 gene poses a great threat to human health by causing the ineffectiveness of the last-resort antibiotic, polymyxins (9). The presence of the mcr-1 gene along with other mcr genes, has been reported from more than 40 countries (10). In Pakistan, the presence of the mcr-1 gene has been detected in Escherichia coli isolated from wildlife, human, poultry, and healthy broiler chickens (11-14). However, scarce data are available about the presence of the mcr-1 gene in other bacterial species in Pakistan.
2. Objectives
The present study aimed to investigate the presence of the mcr-1 gene in K. pneumoniae isolated from urine samples collected from government hospitals located in Peshawar and Islamabad, two major cities of Pakistan.
3. Methods
3.1. Bacterial Isolates
A total of 525 urine samples were collected from three major hospitals, including Khyber Teaching Hospital (KTH) and Combined Military Hospital (CMH) in Peshawar and one major hospital, namely the Pakistan Institute of Medical Sciences (PIMS) Hospital in Islamabad. Sampling was carried out for seven months from January 2017 to July 2017. Urine samples were collected in sterile urine collection bottles and were immediately transferred to the Microbiology laboratories of the respective hospitals. The collected samples were directly streaked on cysteine lactose electrolyte deficient (CLED) media and incubated at 37°C for 24 h. A total of 298 K. pneumoniae isolates were screened through colony morphology, Gram staining, and biochemical tests (15). The isolates were then stored in the Luria-Bertani broth medium with 40% glycerol at -80°C until further processing for the molecular detection of genes. Working cultures were maintained on nutrient agar at 2°C - 8°C for up to four weeks.
3.2. Detection of Extended-Spectrum β-Lactamase Production
Extended-spectrum β-lactamase was detected by the Double Disc Synergy test (DDST) using Mueller-Hinton agar (MHA) following the Clinical Laboratory Standards Institute (CLSI) guidelines (16). The used antibiotic discs were cefotaxime (CTX 30 µg), ceftriaxone (CRO 30 μg), ceftazidime (CAZ 30 µg), cefepime (FEP 30 µg), and amoxicillin + clavulanic acid (AUG 20 µg + 10 µg). Discs containing CTX 30 µg, CRO 30 µg, CAZ 30 µg, and FEP 30 µg were placed around the disc of AUG 20 µg + 10 μg. The observation was made by measuring the distance between the surrounding discs and the extension of the edge of any cephalosporin disc towards the AUG disc (17).
3.3. Detection of Colistin Resistance
The colistin resistance of ESBL-positive isolates was detected by both the agar dilution method and the broth microdilution method. The minimum inhibitory concentrations (MIC) results were interpreted according to the European Committee on Antimicrobial Susceptibility testing (EUCAST) guidelines (18).
3.4. DNA Extraction
The plasmid DNA was extracted by the alkaline lysis method (19) from the colistin-resistant isolates, and the extracts were labeled as KP07, KP09, KP30, and KP31.
3.5. Conventional PCR for mcr-1
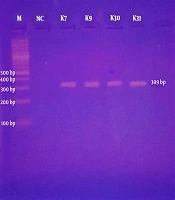
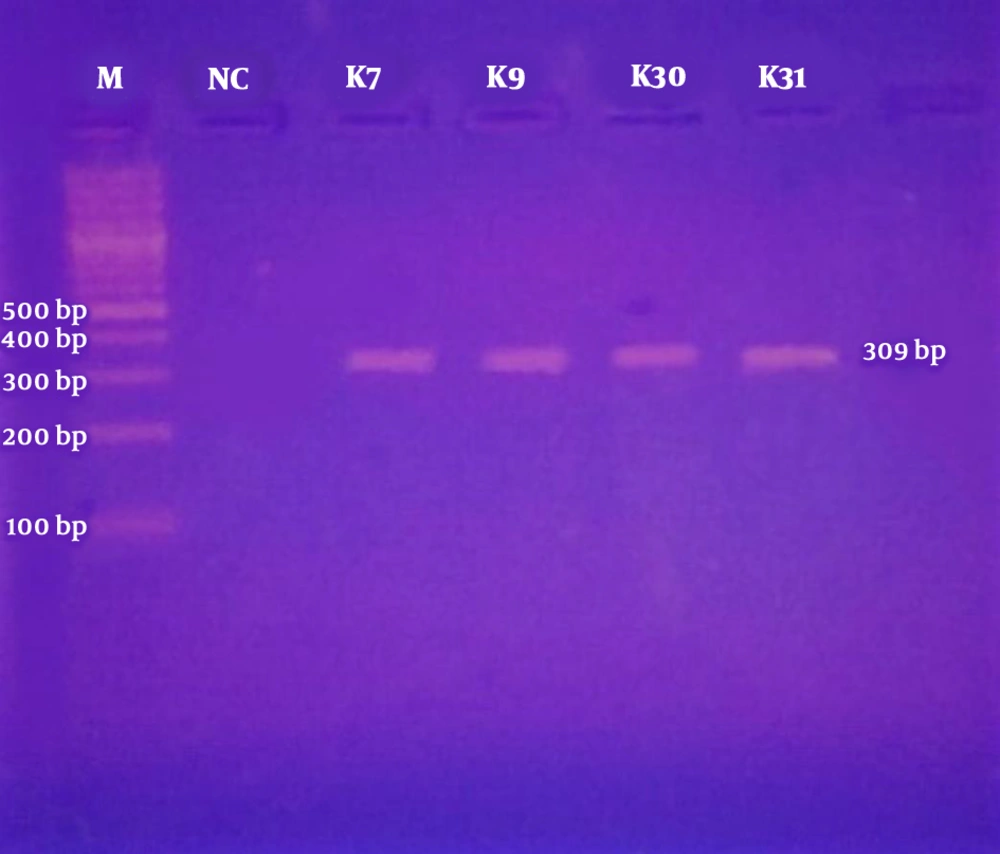
The designed primer sequence was provided to Gene Link for commercial synthesis. The primers used for the amplification of the mcr-1 gene are given in Table 1. The mcr-1 gene was detected in all colistin-resistant isolates by conventional PCR. The product size of mcr-1-positive amplicon was 309 bp, and the PCR cycling conditions were the same as previously described by Liu et al. in 2015. The bands of the expected size were visualized on the 1% agarose gel after electrophoresis at 90 V for 35 min.
| Target Gene | Nucleotide Sequences (5’ → 3’) | Amplicon Size, bp | Source |
|---|---|---|---|
| mcr-1 | MCR1-F: CGGTCAGTCCGTTTGTTC | 309 | (9) |
| MCR1-R: CTTGGTCGGTCTGTAGGG |
Abbreviations: F, forward; R, reverse.
3.6. Sequencing of mcr-1-Positive Amplicons
The PCR products containing mcr-1-positive 309 bp amplicon were confirmed by the Sanger sequencing method using both forward and reverse primers. The attained sequences were compared with the previously published mcr-1 gene sequences in NCBI GenBank (http://www.ncbi.nlm.nih.gov/blast/).
4. Results
In total, 298 clinical isolates were confirmed for K. pneumoniae, including 260 isolates from the PIMS in Islamabad, 22 from the KTH in Peshawar, and 16 from the CMH in Peshawar. The hospital-wise percentage of ESBL-positive K. pneumoniae isolates are presented in Table 2. The ESBL activity was detected in 35 isolates via DDST. Of the 35 isolates, 32 were from the PIMS in Islamabad, two from the KTH in Peshawar, and one from the CMH in Peshawar. Out of 35 ESBL-positive K. pneumoniae isolates, four isolates (three from the PIMS in Islamabad and one from the KTH in Peshawar) showed resistance to colistin (Table 3). Three out of four colistin-resistant isolates (KP07, KP30, and KP31) showed similar MIC results, i.e., 4 mg/L on both broth micro and agar dilution methods. In contrast, one isolate (KP09) showed different MIC results, i.e., 8 mg/L on the broth microdilution method and 4 mg/L on the agar dilution method.
The plasmid DNA was extracted from these four isolates for use as a template in conventional PCR. In colistin-resistant isolates, the mcr-1-specific primers amplified the desired region of 309 bp (Table 3); they were visualized on the 1% agarose gel, as shown in Figure 1. The demographic data of patients and MIC distribution of the four mcr-1-positive isolates are presented in Table 4. The resulted mcr-1 amplicons of 309 bp length were confirmed by Sanger sequencing. The sequence analysis of all the query sequences confirmed 99% sequence similarity with the mcr-1 resistance gene of E. coli (GenBank accession number: LC427672.1). The sequence of K. pneumoniae strain was submitted to NCBI GenBank (GenBank accession number MK340993).
| S. Number | Hospitals | Collected Isolates | ESBL-Positive |
|---|---|---|---|
| 1 | PIMS Islamabad | 260 (87.24) | 32 (12.30) |
| 2 | KTH Peshawar | 22 (7.38) | 2 (9.09) |
| 3 | CMH Peshawar | 16 (5.36) | 1 (6.25) |
| Total isolates | 298 | 35 (11.7) |
Abbreviations: CMH, Combined Military Hospital; ESBL, extended-spectrum β-lactamase; KTH, Khyber Teaching Hospital; PIMS, Pakistan Institute of Medical Sciences.
aValues are expressed as No. (%).
| Bacterial spp. | ESBL Producers | Colistin-Resistant | Isolates Harboring the mcr-1 Gene |
|---|---|---|---|
| Klebsiella pneumoniae | 35 (11.7% of the total isolates) | 4 (11.42% of the ESBL producers) | 4 (100% of the colistin-resistant isolates) |
Abbreviation: ESBL, extended-spectrum β-lactamase.
aValues are expressed as No. (%).
Ethidium bromide-stained 1% agarose gel showing PCR-amplified mcr-1 gene fragments with specific primers. Lane M, represents 100 bp DNA marker (Bio-Rad); Lane NC, is a negative control; Lanes K7, K9, K30, and K31, are Klebsiella pneumoniae isolates showing the expected bands of 309 bp of mcr-1 gene.
| Isolate ID | Patients | Sample | MIC of Colistin, mg/L | Mcr-1 | ||
|---|---|---|---|---|---|---|
| Gender | Age, y | Agar Dilution Method | Broth Micro-Dilution Method | |||
| KP07 | Female | 36 | Urine | 4 | 4 | + |
| KP09 | Female | 45 | Urine | 4 | 8 | + |
| KP30 | Male | 40 | Urine | 4 | 4 | + |
| KP31 | Female | 52 | Urine | 4 | 4 | + |
5. Discussion
This study identified, for the first time, mcr-1 harboring K. pneumoniae in human urine samples collected in Pakistan. Additionally, these isolates were also ESBL-positive. Colistin is the last resort antibiotic available to date against MDR bacteria, particularly ESBL and carbapenem-resistant Enterobacteriaceae (CRE) harboring the NDM 1 and KPC 2 genes (20). After the increased emergence of CRE, the use of colistin increased in both human and animal medicine (21). This resulted in the emergence of a new plasmid-mediated resistance gene named mcr-1. Its presence on the plasmid is a matter of concern due to the plasmid’s ability of horizontal transfer via bacterial conjugation (22).
In the present study, the prevalence of ESBL-producing K. pneumoniae from urine samples was 11.7%, which is in agreement with a study from Sri Lanka conducted by Fernando et al. (23) in 2017 reporting 13.8% of the K. pneumoniae isolates as ESBL producers. A study from Lalitpur, Nepal, conducted by Shakya et al. (24) in 2017 reported 17.64% of the total investigated strains of K. pneumoniae as ESBL producers. In another study reported by Ahmed et al. (25) from Pakistan, 24.5% of the K. pneumoniae isolates were shown to be ESBL-positive, which is higher than the percentage in the current study. Batool et al. (26) from Pakistan in 2016 reported 34% of the K. pneumoniae isolates as ESBL-positive among 97 Gram-negative rods. Another study from Pakistan conducted by Ejaz (27) reported 71.75% of the K. pneumoniae isolates as ESBL producers. The differences in the prevalence could be due to different techniques used for the phenotypic identification of ESBL-producing isolates (28) and/or due to differences in geographical regions (29).
Colistin resistance was detected phenotypically using both agar and broth dilution methods, the results of which were the same in three out of four isolates while in one isolate (KP09), the MIC values were different, i.e., 8 mg/L and 4 mg/L, respectively. However, it did not affect the resistance breakpoint for colistin, which is 2 mg/L according to the EUCAST guidelines (30).
The presence of the mcr-1 gene has been reported in K. pneumoniae from different countries. A study from South Africa by Newton-Foot et al. (31) reported the mcr-1 gene in five isolates of K. pneumoniae from humans during seven months of the study. Another study from Laos by Rolain et al. (32) reported four isolates of K. pneumoniae harboring the mcr-1 gene, which is similar to our findings of the mcr-1 gene in four isolates. However, the isolates in Rolain et al. (32) study were not ESBL-positive. The emergence of antibiotic resistance genes in urine samples is a critical issue because of the poor drainage system in underdeveloped countries like Pakistan (33). Thus, resistant isolates can come in direct contact with people in drinking water and other food items (34).
In Pakistan, there is excessive use of colistin alone or in combination with other antibiotics for curing colibacillosis and clostridial enteritis in poultry (35). This increased use of colistin is directly related to the emergence of the mcr genes and resistance to colistin in bacterial isolates from poultry (14) and spread to humans through the food web (36, 37). If the consumption of colistin continues to increase at the same pace, we will enter the post-antibiotic era with the widespread emerging resistance to colistin. It is projected that the utility of new antimicrobial agents increases by up to 67% by 2030 (38). The guidelines for the use of antibiotics in animal husbandry and human wellbeing should be applied globally to minimize the risk of antimicrobial resistance.
5.1. Conclusions
Our study concludes that the mcr-1 gene exists in ESBL-producing K. pneumoniae in our locality. It is an alarming issue as mcr-1 in K. pneumoniae can be easily transferred to other bacterial species via horizontal gene transfer. Thus, urgent measures should be adopted to overcome the inappropriate use of colistin-containing formulations with the hope of preventing further spread of resistance to this antibiotic.