1. Background
Staphylococcus aureus is a commensal bacterium that usually colonizes the skin and mucous membranes and causes a wide variety of diseases, including soft-tissue and bloodstream infections and life-threatening sepsis (1, 2). Peptidoglycan biosynthesis in the cell wall of Gram-positive bacteria starts with membrane-bound enzymes called penicillin-binding proteins (e.g., PBP2). Due to the acquisition of mecA in certain strains, a new protein called PBP2a is encoded. This protein cannot attach to beta-lactam antibiotics, although it retains the enzymatic activity; thus, these strains are called methicillin-resistant S. aureus (MRSA) (3).
In recent years, the frequency of MRSA has increased to 60% of the clinical isolates of S. aureus (4, 5). Some clinical mecA and PBP2 positive strains of S. aureus exhibit phenotypic susceptibility to oxacillin. It has been suggested that such isolates can be classified as a new group of MRSA called oxacillin-susceptible methicillin-resistant S. aureus (OS-MRSA). They may be incorrectly classified as methicillin-sensitive S. aureus (MSSA) just because they show phenotypic susceptibility to oxacillin (6, 7). It is estimated that the prevalence of MRSA and the mortality rate of S. aureus bacteremia in many countries have raised to more than 49% and 50%, respectively (8, 9). Recently, the epidemiology of MRSA has changed, and several nosocomial strains have produced community-acquired infections, named community-onset hospital-acquired infections (CO-HA MRSA), or vice versa (HO-CA MRSA) (10, 11).
The mecA gene is carried by the cassette chromosome mecA (SCCmec). Eleven mecA cassettes (SCCmec) are recognized as S. aureus (12, 13). The majority of the prevalent MRSA strains in the community are SCCmec type IV, while types I, II, and III are associated with hospital-acquired MRSA infections (5, 14). The routine detection methods of MRSA, like culture and PCR, have several strengths and weaknesses. The disadvantages of these techniques include the high cost of the instruments, and more importantly, the inability to apply as a point of care test (15, 16). Thus, the development of a low-cost and fast method with high sensitivity and specificity is of utmost importance (17, 18).
As an alternative method for the diagnosis of MRSA, loop-mediated isothermal amplification (LAMP) was developed by Notomi et al. (19) in 2000. This method uses the Bst DNA polymerase that has the ability of strand displacement and yields large fragments of DNA by the auto-cycling reaction. In addition, the LAMP reaction is performed at 60°C - 65°C and is carried out for 60 to 90 min (20, 21). This method involves six primers especially designed to recognize a total of six distinct sequences on the target DNA. The LAMP method has been used to identify many bacteria, such as Helicobacter pylori and Arcobacter, and many parasites, fungi, and viruses (1, 6, 21, 22).
2. Objectives
Concerning the high prevalence of the infections caused by MRSA strains, especially in immune-compromised patients, and the importance of this bacterium in nosocomial infections, we attempted to use LAMP for the diagnosis of MRSA in clinical settings.
3. Methods
3.1. Bacterial DNA
Serial dilutions of DNA were prepared from a confirmed clinical isolate of S. aureus. Bacterial DNA was extracted from a 24-h cultured organism using the CinnaPure-DNA kit following the manufacturer’s protocol.
3.2. Clinical Sample
During 2013 to 2014, 53 suspected clinical samples of MRSA and MSSA isolated at the Microbiology Laboratory of Imam Reza Hospital, Kermanshah, Iran, were re-identified to the species level using standard biochemical tests like Gram staining, catalase, DNase, coagulase, oxidase, and sensitivity to bacitracin (23).
3.3. Antimicrobial Susceptibility Testing
The disk diffusion and mico-dilution broth methods were carried out according to the standard methods using cefoxitin and oxacillin disks (MAS diagnosis) and oxacillin powder (Sigma), respectively. The in vitro antimicrobial susceptibility testing for MRSA was performed using the Kirby-Bauer disk diffusion method (Clinical Laboratory and Standard Institute, CLSI). The MIC of oxacillin was determined using serial dilutions from 0.5 to 1,024 µg/mL of the antibiotic (CLSI, 2016) (24).
3.4. PCR and LAMP Primer Design
The sets of forward and reverse PCR primers for 16sRNA, mecA, and spa genes were designed manually and confirmed by the online Primer3 software (Table 1). The LAMP primers including outer forward primer (F3), outer backward primer (B3), forward inner primer (FIP), and backward inner primer (BIP) targeting the spa and mecA genes were designed by online LAMP primer design software Primer Explorer V4 (Table 2).
| Target Gene | Primer (5’ → 3’) | Size of Amplicon |
|---|---|---|
| spa | F: CTAATAACGCTGCACCTAAG | 309 |
| R: GCAAAGAAGATGGCAACAAAC | ||
| mecA | F: ACCACTTCATATCTTGTAACG | 160 |
| R: AGATTACAACTTCACCAGGTTC | ||
| 16srRNA | F: GCAAGCGTTATCCGGATTT | 597 |
| R: CTTAATGATGGCAACTAAGC |
| Target Gene | Primer (5’ → 3’) |
|---|---|
| spaF3 | TTATAGTTCGCGACGACG |
| spaB3 | TGCAGATAACAAATTAGCTGAT |
| spaFIP | CCAGAAACTGGTGAAGAAAATCCATGAATTCCTAAGGCTAATGATAATCCACC |
| spaBIP | AATGCTTGAGCTTTGTTAGCATCT-GAATTC-AACATGATCAAACCTGGTCA |
| spaLF | TCATCGGTACAACTGTA |
| spaBF | GCATGGTTTGCTGGTTGC |
| mecAF3 | AGTTCTTTAGCGATTGCTTTA |
| mecAB3 | CATTGATCGCAACGTTCAA |
| mecAFIP | CGAAACAATGTGGAATTGGCCAA-GAATTC-ATTCTTTGGAACGATGCCTAT |
| mecABIP | GCTTTGGTCTTTCTGCATTCCTG-GAATTC-AGAAGATGGTATGTGGAAGT |
| mecALF | ACAGGAACAGCATATGAG |
| mecABF | ATGACGCTATGATCCCAA |
3.5. Molecular Methods
3.5.1. Multiplex PCR
First, the uniplex PCR was set up for 16sRNA, spa, and mecA separately. The PCR mixtures contained 1.5 µL 10x PCR buffer, 1.5 mmol MgCl2, 0.2 mmol dNTP, 1 µmol of each of the designed primers, 1 U Taq DNA polymerase, 2.0 µL template DNA, and 8.55 µL distilled H2O to a final volume of 15.0 µL. The optimum thermal cycling program for all the gene targets was set as follows: initial denaturation at 95°C for 5 min, followed by 34 cycles of denaturation at 95°C for 30 s, annealing at 53°C for 30 s, extension at 72°C for one minute, and a final extension at 72°C for 5 min. As we aimed to identify the spa and mecA genes simultaneously, the designed program was used to amplify the genes at the same time in a multiplex reaction. Therefore, the PCR mixture was prepared as follows: 2 µL 10× PCR buffer, 0.6 µL MgCl2, 0.5 µL dNTP, 2 µL each primer, 0.3 µL Taq DNA polymerase, 2.0 µL template DNA, and 5.6 µL H2O to a final volume of 15.0 µL. The program was set as mentioned before, except that the annealing temperature was set at 54°C for both genes. Finally, the PCR products were detected using electrophoresis on a 1.5% agarose gel using ethidium bromide staining.
3.5.2. Multiplex LAMP
The uniplex LAMP was set up for the spa and mecA genes before designing multiplex LAMP. The LAMP mixture was set as follows: 2 µL 10× buffer, 2.5 µL MgSO4, 5 µL dNTP, 4 µL Betine, 0.4 µL of each of FIP and BIP primers, 0.5 µL of each of LF and LB primers, 2 µL template DNA, 1.1 µL Bst polymerase, and 4.3 µL H2O to a final volume of 25 µL. This program was set as follows: initial denaturation at 92°C for 5 min, 60.5°C for 40 min, and a final extension at 92°C for 3 min. The multiplex LAMP reaction for the spa and mecA genes was carried out on a scale of 30 µL mixture containing 6 µL 10× LAMP buffer (1.5×), 3.2 mmol (8 µL) dNTP, 0.8 µmol betaine, and 8 mmol MgSO4. For the mecA gene, 1.2 µmol/L of primers FIP and BIP, 0.16 µmol/L of primers F3 and B3, and 0.4 µmol/L of primers LF and LB were applied. For the spa gene, 1.6 µmol/L of primers FIP and BIP, 0.2 µmol/L of primer F3, 0.16 µmol/L of primer B3, and 0.8 µmol/L of primers LF and LB were mixed. Besides, 2 µL of template DNA and 4.2 µL of sterile deionized water was used for all the mixtures and were incubated at 92°C for 5 min. After this step, 1.7 µL of Bst DNA polymerase was added and incubated at 60.5°C for 80 min, heated to 92°C for 3 min, and finally cooled at 4°C for 5 min to terminate the reaction. For further confirmation, the products were also detected using electrophoresis on a 1.5% agarose gel using ethidium bromide staining.
3.6. Analytical Sensitivity of PCR and LAMP
The analytical sensitivity of LAMP and PCR was determined Using nanodrop instrument, the amount of DNA in the first concentration was determined as 83.3 mg/mg. Then, different concentrations of pure DNA were prepared by serial dilutions.
3.7. Analytical Specificity of LAMP and PCR
To determine the analytical specificity of designed LAMP and PCR for the mecA and spa genes, the total genomic DNA of methicillin-sensitive S. epidermidis, Klebsiella, Acinetobacter, Escherichia coli, Citrobacter freundii, Enterococcus faecalis, Micrococcus, S. agalactiae, S. saprophyticus, and Enterobacter was tested by the designed methods.
3.8. Clinical Sensitivity and Specificity
Fifty-three isolates of suspected S. aureus were determined by biochemical methods and confirmed by the amplification of the 16S rRNA gene. Resistance to methicillin was surveyed by disk diffusion using cefoxitin and oxacillin disks; the MIC of oxacillin was determined, as well. To determine the clinical sensitivity and specificity of PCR and LAMP, these tests were compared with Gold standards, including culture, antibiogram, and MIC.
3.9. Statistical Analysts
The data from LAMP and PCR methods were analyzed using SPSS version 21. The agreement between the two methods was analyzed by the kappa coefficient test.
4. Results
4.1. Analytical sensitivity of LAMP and PCR
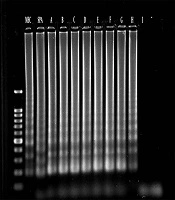
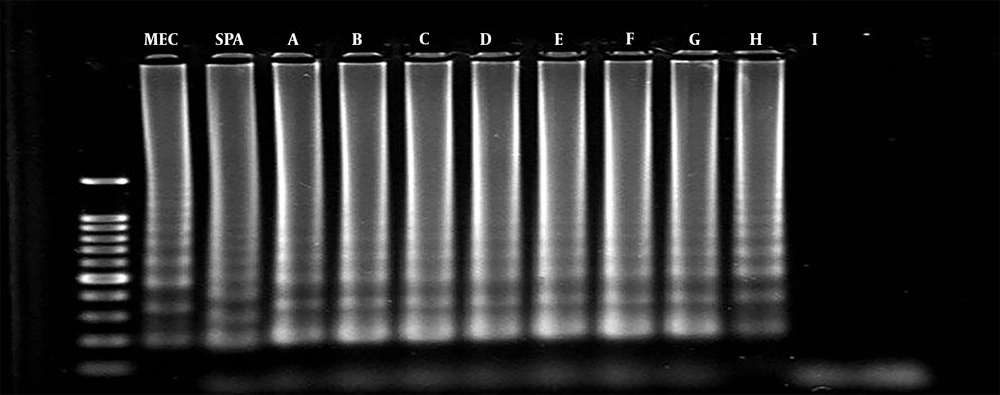
The analytical sensitivity of multiplex PCR and multiplex LAMP was determined using serial dilutions of S. aureus DNA samples as the target DNA. The PCR could detect the mecA and spa genes at 1 fg/mL and 0.01 pg/mL of bacterial DNA, which were equal to 35 and 350 copy numbers of bacterial DNA, respectively. Similarly, multiplex LAMP detected the spa and mecA genes at 0.1 fg/mL and 1 fg/mL of bacterial DNA, which were equal to 3.5 and 35 copy numbers of bacterial DNA, respectively (Figures 1 and 2).
4.2. Analytical Specificity of LAMP and PCR
Multiplex LAMP and multiplex PCR were carried out on methicillin-sensitive S. epidermidis, Klebsiella sp., Acinetobacter sp., E. coli, Citrobacter sp., E. faecalis, Micrococcus sp., S. agalactiae, S. saprophyticus, and Enterobacter sp. Amplification was not observed in both tests, which showed a 100% analytical specificity of the tests.
4.3. Clinical Sensitivity and Specificity of Multiplex PCR and Multiplex LAMP
In this study, 53 clinical S. aureus suspected isolates were re-identified by standard biochemical tests, of which 49 isolates were S. aureus and four cases were non-aureus. According to the results of MIC of oxacillin sodium salt and antibiogram tests, 37 isolates were MRSA (oxacillin 4 > MIC µg/mL) and 16 isolates (MIC < 2 μg/mL) were MSSA. According to multiplex PCR, 49 isolates harbored spa, and the sensitivity and specificity were 100% compared to culture as the Gold standard.
Of the 49 isolates, 40 isolates harbored the mecA gene and showed as MRSA. Compared to the results of MIC and disk diffusion as Gold standards with a 95% confidence interval, the sensitivity, specificity, positive predictive value, and negative predictive value of multiplex PCR were 97.3%, 75%, 90%, and 31.92%, respectively. In multiplex LAMP, the sensitivity, specificity, positive predictive value, and negative predictive value of the test for the assessment of the spa gene were 100%, 100%, 100%, and 100%, respectively. Similarly, the sensitivity, specificity, positive predictive value, and negative predictive value of the test for the assessment of the mecA gene were 100%, 75%, 90.2%, and 100%, respectively.
4.4. Comparison of Multiplex LAMP and Multiplex PCR in Clinical Samples
As many as 40 strains harboring the mecA gene were observed in both methods, while 12 strains did not have the gene. Multiplex LAMP detected the mecA gene in one mecA multiplex PCR-negative isolate. Altogether, the sensitivity, specificity, positive predictive value, and negative predictive value of LAMP compared to PCR were 97.96%, 100%, 100%, and 92.31%, respectively. The kappa coefficient was calculated as 0.948.
5. Discussion
Methicillin-resistant S. aureus is a human pathogen that acts as both community and hospital-acquired infections worldwide. The increased MRSA frequency among health care centers has become a major problem in public health and infection control programs (14). Therefore, the rapid and accurate diagnosis of MRSA helps prevent the spread of nosocomial infections (25). Various methods like culture, PCR, MIC determination, and antibiogram are applicable for the detection of MRSA, although all of the mentioned methods suffer from some drawbacks. Molecular methods like PCR and real-time PCR are considered as methods of choice, as they are sensitive, accurate, and less time-consuming in comparison with culture and biochemical methods (26, 27).
Loop-mediated isothermal amplification is used for the rapid detection of DNA and RNA molecules. This technique, unlike PCR, does not need post-amplification procedures to detect the product and is advantageous compared to real-time PCR as it does not need expensive instruments (26, 27).
According to our experiment, the multiplex LAMP could detect the sequences of staphylococcal mecA and spa genes significantly more accurately than did multiplex PCR. This deduction was quite predictable considering similar publications, although the difference is promising. The calculated sensitivity of the multiplex LAMP in our study was much higher than that reported in similar experiments by Tie et al. and Su et al. (4, 17, 20). In a similar study conducted on the species of Helicobacter, higher percentages were reported for the sensitivity and specificity of multiplex LAMP than those of other methods such as multiplex PCR (21, 22).
Some of the features of the LAMP method include higher specificity and sensitivity and less time, labor, and equipment consuming than PCR. Moreover, since a large quantity of products is produced in the amplification by LAMP, the final amplicon detection can be performed without the need for gel electrophoresis equipment and it also allows for easy visualization by the naked eye (28). Typically, six primers (three pairs of primers) are used for the detection of about 100 bp of target DNA while, in PCR, two primers are attached to 40 bp. In this study, LAMP could detect the genes in clinical specimens in 188 minutes.
5.1. Conclusions
We introduced the multiplex loop-mediated isothermal Amplification as an alternative method for the diagnosis of methicillin-resistant S. aureus. This method has several advantages over PCR and culture, consisting of higher sensitivity and specificity.