1. Background
Infections are principal restraints in the culture of many aquatic species, because of economic and social expansions in aquaculture production and trade (1). Bacteria are one of the most well-known fish pathogenic agents (2). The most common bacterial agents related to fish diseases in the marine/brackish water and freshwater environments are vibriosis and motile aeromonads, respectively (3). Aeromonas spp. are Gram-negative bacteria that can live in environments such as groundwater or chlorinated drinking water (4). This genus contains different species among which, Aeromonas veronii biovar veronii, A. caviae, A. hydrophila, and A. salmonicida are well known (5). Aeromonas spp. are ubiquitous bacteria and their isolation is expected to occur from extensively varied humid environments, as well as sublime, stagnant, and freshwaters, sewage, fish with or without clinical signs, and marine and animal products or feces (6).
Some of the various Aeromonas species are isolated from various human infections; thus, their identification is essential (5). The prevalence of these species in aquatic environments, and similarly in food, is considered a potential health risk for humans (7). Two major categories of human illnesses attributed to Aeromonas species include acute gastroenteritis in both pediatric and adult populations and disseminated disease (e.g., bacteremia) in individuals with underlying hematologic malignancies or hepatic dysfunctions. There are multiple Aeromonas virulence factors including adhesins, S/A layer, lipopolysaccharides, motility, siderophores, biofilm-producing ability, and the mass of exoenzymes as extracellular products with biological activities such as enterotoxin, cytotoxins, aerolysin, protease, and lipases (8).
Biofilm is a bacterial structure with an important role in worsening human and animal infections. In fact, in this community, the penetration of antibiotics and native immunity effects is limited and the probability of antibiotic resistance gene transfer among bacteria relative to planktonic cells is high (9) resistance could be reflection of the generated nutrient environment in biofilm structure, direct modification of antibiotic action through the presence of extracellular polymers, the development of attachment/biofilm specific phenotype, antibiotic modification enzyme or foster the plasmid exchange in biofilm structure.
Oluwasusi et al. compared bacterial counts and antibiotic resistance patterns of different isolated bacteria with biofilm-producing ability in water from boreholes and wells. Based on the results, the highest bacterial counts and most resistance to antibiotics were detected in borehole isolates. The findings of this study suggest the high prevalence of multiple antibiotic resistance indices, indicating high contamination in areas where antibiotics were used (10). Moreover, Onyebuchi et al., Oluyege et al., and Segev-Zarko et al. conducted studied in this context and provided similar multiple antibiotic resistance patterns in different isolated biofilm-producing bacteria with the same origin (water) (11-13). Aeromonas can colonize on biotic surfaces such as epithelial cell lines or vegetable salads and abiotic surfaces such as plastic, glass, and stainless steel. Biofilm production needs to be realized in varied condition including food processing systems, gastrointestinal tracts, and water distribution systems (14).
Biofilm structure could serve a permanent origin of cross-contamination in the course of aquatic food preparation (15) and is hazardous for public health via the consumption of contaminated foods and direct contact with infected animals (16, 17). Biofilm formation is reported in several Gram-positive bacteria including Staphylococcus spp., Streptococcus spp., and Pseudomonas spp. and Gram-negative bacteria such as Vibrio spp., Klebsiella spp., and Escherichia coli. Numerous studies permitted the documentation of several essential factors complicated in biofilm formation. Prerequisite factors include essential components involved in attachment and dispersion such as membranous proteins and polysaccharides, lateral or polar flagella, interconnecting cell components, as well as culture media composition, temperature, pH, oxygen accessibility, and some genetic features such as plasmids (18). Thus far, little is known about the physiological alterations that arise in biofilm formation in the case of Aeromonas species. It has been shown that polar and lateral flagella can increase biofilm formation (19). Moreover, bacterial biofilm formation is favored under most nutrient-sufficient environments.
2. Objectives
It is critical to identify Aeromonas spp. with biofilm-producing ability since they are predominant pathogens in aquaculture, as well as in humans as opportunistic pathogens. Hence, the current study aimed at emphasizing the pathogenic strains of A. hydrophila with biofilm formation ability using antibiograms with common antibiotics in human and marine cultures.
3. Methods
3.1. Isolation of Pathogenic Aeromonas hydrophila
Aeromonas hydrophila studied in the current study was isolated from 120 carp fishes with clinical signs of septicemia in several farms of Khuzestan province, Iran. For this purpose, a culture was prepared from the fish kidney in blood agar and incubated at 37ºC for 24 h. After the selection of suspicious colonies, they were propagated by renewed culture on blood agar and then identified by catalase, oxidase, Gram staining, and biochemical tests including triple sugar iron agar, Indole, urease, Simmon citrate, lipase test, motility, lysine/ornithine decarboxylase, and arginine dihydrolysis (20).
3.2. Polymerase Chain Reaction
For molecular identification, the presence of A. hydrophila genus- and species-specific genes was determined using PCR. First, DNAs were extracted from bacterial suspensions in TE buffer (Tris-EDTA) containing 2-mercaptoethanol (2% v/v) by boiling. The supernatants were then collected as a source of DNA after centrifugation. The designed genus- and species-specific primers for the 16S rRNA gene are demonstrated in Table 1. Duplex-PCR was performed in 25 µL total volume, containing 10 pmol/µL of each primer (1 µL) and 3 µL of extracted DNA in 12.5 µL of PCR master mix 2X (Ampliqon, Denmark), in addition to 5.5 µL of nuclease-free water. The PCR protocol was as follows: one cycle at 94ºC for 4 min, 30 cycles at 94ºC for 30 s, 60ºC for 30 s, and 72ºC for 30 s, followed by a final extension at 72ºC for 7 min in a thermal cycler (Eppendorf, Germany). Aeromonas hydrophila (ATCC 7966) and nuclease-free water were used as positive and negative controls, respectively. The PCR products were visualized by electrophoresis on the 1% agarose gel (Max Pure, Spain) and stained with safe stain (1×) (Sinaclon, Iran) using a UV transilluminator (UV tech, Germany) (21, 22). The demonstration of the 599 bp segment in each sample indicated Aeromonas genus positivity and of the 685 bp segment indicated hydrophila species positivity.
3.3. Antibiotic Susceptibility Testing
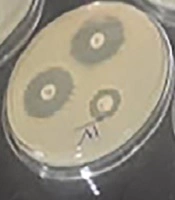
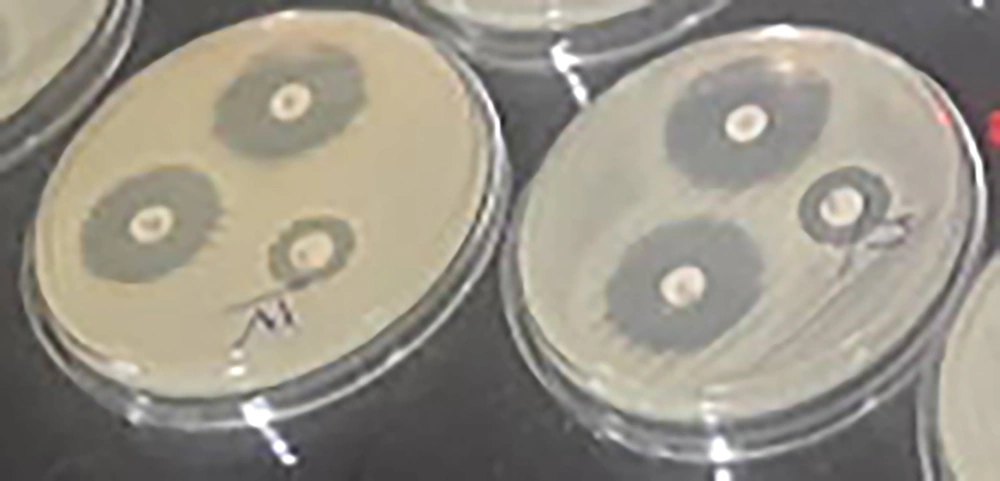
In the current study, susceptibility to several antibiotics was determined by the disc diffusion technique using standard antibiotic discs (PadtanTeb) (23). The bacterial inoculum of each studied strain was adjusted to a certain concentration equal to 0.5 McFarland’s turbidity (approximately 2 × 108 CFU/mL) by the sterile cotton-tipped applicator and inoculated onto the entire surface of Mueller-Hinton Agar (MHA) medium. Then, several standard antibiotic discs such as ciprofloxacin (CP) (5 mg), clindamycin (CC) (10 mg), vancomycin (V) (30 mg), tetracycline (T) (30 mg), sulfamethoxazole-trimethoprim (SXT) (23.7 + 1.25 mg), oxytetracycline (TE) (30 mg), amoxicillin (AMK) (10 mg), and streptomycin (S) (15 mg) were placed on the prepared cultured medium. After aerobic incubation for 24 - 48 h at 25ºC, the inhibitory zone of each disc was measured. The results were interpreted based on the CLSI criteria to express the susceptibility or resistance of each isolate to studied antibiotics (24). In this step, A. hydrophila standard strain (ATCC7966) was used as a positive control.
3.4. Biofilm Assay
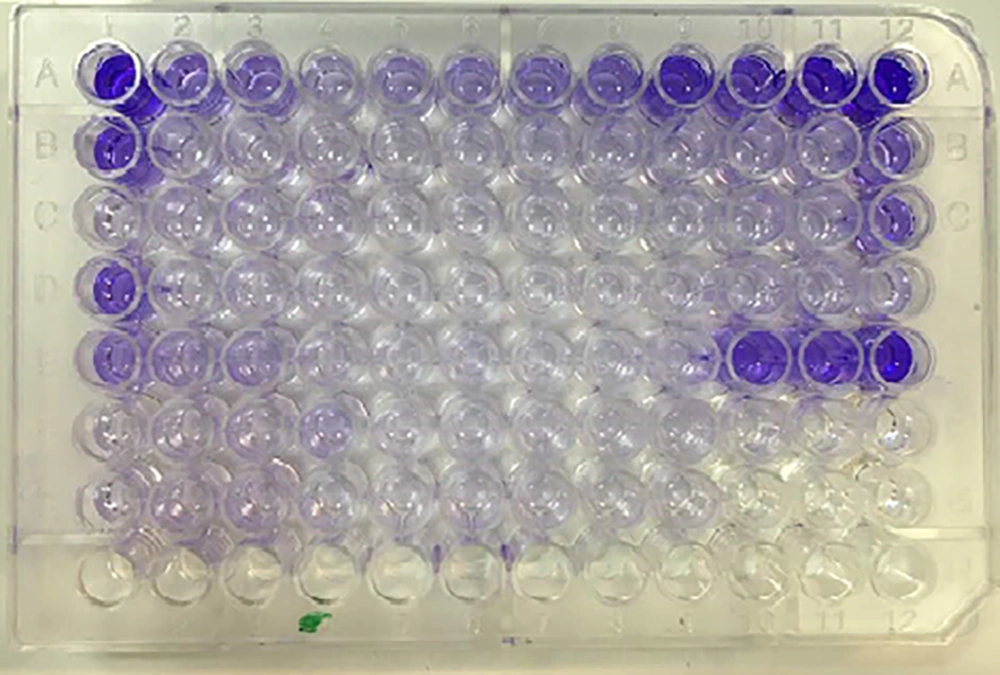
Biofilm formation was evaluated by the quantitative microtiter plate crystal violet method as described by Stepanovic et al., with some modifications. First, in the primary study (data not shown), four culture media (tryptic soy broth, peptone water, Luria-Bertani, and brain-heart infusion broth) were evaluated to determine the best culture medium to encourage biofilm production using three standard strains. After the selection of Trypticase Soy Broth (TSB) (Merck, Germany) as the best culture medium, 10 mL of TSB was inoculated by an adequate amount of overnight culture of each isolate to adjust to an optical density (OD) of 0.8 at 600 nm. Then, it was spread on a polystyrene microtiter plate (Max-well) by a 100 µL single-channel sampler (Brand, Germany) and incubated for 24, 48, 72, and 96 h at 37ºC and 28ºC without shaking. Non-inoculated supplemented TSB was used as the negative control in triplicate. Biofilm production was analyzed at different incubation times (24, 48, 72, and 96 h) at 28ºC and 37ºC. After the selection of the best conditions and culture of strains in it, the plate was washed by sterile normal saline in triplicate.
The plate was dried at room temperature and 200 µL of methanol per each well was added for 15 min. After plate aspiration and drying at room temperature, the plate was stained with 200 µL of 2% solution of Hucker’s crystal violet. Washing and drying were carried out again after 5 min; then, 200 µL of discoloring solution per each well (ethanol-acetone) was added and after 15 min, the absorbance was measured using an ELISA (enzyme-linked immunosorbent assay) plate reader (Biotek SX2, USA) at 600 nm. The cutoff OD was defined as the mean OD of negative controls. Each sample was tested in triplicate and in three repeats. Biofilm-positive control was A. hydrophila standard strain (ATCC7966). The OD of each strain was obtained by the arithmetic mean of the absorbance of three wells and this value was compared with the mean absorbance of negative controls (ODnc). The following classification was used to determine biofilm formation: no biofilm formation (ODs < ODnc), weak biofilm formation (ODnc < ODs < 2.ODnc), moderate biofilm formation (2.ODnc < ODs < 4.ODnc), and strong biofilm formation (4.ODnc < ODs) (23, 25). In the end, biofilm-producing ability and antibiotic susceptibility patterns of the strains were compared by the Fisher exact test.
3.5. Statistical Analysis
Fisher’s exact test was used to compare biofilm-producing ability and susceptibility to each antibiotic.
4. Results
Based on biochemical tests and PCR assays, 19 strains of A. hydrophila were isolated from clinical samples. All the strains were Gram-negative, rod-shaped bacteria with positive catalase and oxidase reactions. Most of the isolates were detected as glucose fermenter (97%) and indole positive (95%) and all of them were positive in motility, arginine dihydrolysis, lysine decarboxylase, and lipase tests (100%). In all of them, the hydrolysis of urea and decarboxylation of ornithine were negative and citrate was positive in 85% of the isolates. Moreover, β-hemolysis varied in different strains (Table 2). By duplex-PCR, all the suspected isolates were the carriers of the 16SrRNA genus-specific gene (599 bp) and 16SrRNA species-specific gene (685 bp) in gel electrophoresis (Figure 1). The antibiogram test results of the studied strains and standard A. hydrophila strain (ATCC 7966) are shown in Table 3. The most commonly detected resistance was to clindamycin (18/20) and vancomycin (18/20). Susceptibility to ciprofloxacin, trimethoprim-sulfamethoxazole, and tetracycline was detected in all the A. hydrophila strains. Moderate resistance/susceptibility to streptomycin, oxytetracycline, tetracycline, amoxicillin, and clindamycin was reported in 10 strains, six strains, five strains, one strain, and one strain, respectively (Figure 2 and Table 3).
| Test | Biochemical Test | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Glucose | Cit. | Urea | Indole | Motility | Lipase | LDC | ODC | ADH | |
| Positivity % | 97 | 85 | 0 | 95 | 100 | 100 | 100 | 0 | 100 |
| Antibiotic Type | Susceptible (%) (N = 20) | Moderate (%) (N = 20) | Resistance (%) (N = 20) |
|---|---|---|---|
| Clindamycin | 1 | 1 | 18 |
| Vancomycin | 2 | - | 18 |
| Tetracycline | 15 | 5 | - |
| Oxytetracycline | 13 | 6 | 1 |
| Trimethoprim- sulfamethoxazole | 20 | - | - |
| Ciprofloxacin | 20 | - | - |
| Amoxicillin | 14 | 1 | 5 |
| Streptomycin | 8 | 10 | 2 |
The biofilm-producing ability in the studied strains was evaluated by the microtiter plate crystal violet assay and comparing the mean OD of three wells of each sample (ODs) with the mean OD of three wells of negative control (ODnc = 0.044). Based on the results, the TSB medium with 94 h incubation at 28ºC was the best condition for biofilm production. In this condition, four (21%) strains in addition to the standard strain were detected as strong biofilm producers (0.176 < ODs) and 15 (79%) strains as moderate biofilm producers (0.088 < ODs < 0.176). The standard strain as the positive control was evaluated as the strong producer (OD = 0.611) of biofilm (Figure 3). The correlation between biofilm-producing ability and antibiotic susceptibility pattern of each strain is demonstrated in Table 4.
| Biofilm Ability | Antibiotic Susceptibility | |||||||
|---|---|---|---|---|---|---|---|---|
| CC | V | TE | T | AMX | S | Cp | SXT | |
| Strong (n = 4) | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 4 |
| Moderate (n = 15) | 1 | 2 | 13 | 15 | 14 | 8 | 15 | 15 |
| Positive control | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Total | 1 | 2 | 13 | 15 | 14 | 8 | 20 | 20 |
Isolates with strong producing ability were resistant to 75% of the studied antibiotics. The most susceptibility in all strong biofilm-producing strains was reported to ciprofloxacin and sulfamethoxazole-trimethoprim. In moderate biofilm-producing ability strains (Table 3), the most susceptibility was reported against tetracycline, ciprofloxacin, and sulfamethoxazole-trimethoprim (100%), followed by amoxicillin (93.3%) and oxytetracycline (86.6%). The least susceptibility of these strains was detected against clindamycin (6.6%), vancomycin (13.3%), and streptomycin (53.3%) (Table 4). Base on the Fisher exact test, the correlation between antibiotic resistance and biofilm-producing ability in all strains was not significant (P > 0.05), but a significant correlation was detected between the level of biofilm-producing ability and resistance to some antibiotics (P < 0.01). In fact, there were significant differences between strong and moderate biofilm-producing strains in the frequency of resistance to oxytetracycline (P < 0.01), tetracycline (P < 0.001), and amoxicillin (P < 0.001) but not in the frequency of resistance to clindamycin, vancomycin, streptomycin, ciprofloxacin, and sulfamethoxazole-trimethoprim (P > 0.05).
5. Discussion
Biofilms are the communities of microorganisms in the environment or body that protect individual cells from unsuitable conditions (26). In the biofilm structure, virulence and pathogenicity of bacteria are increased since bacterial surface antigens could be hidden in the biofilm that is a non-penetrative structure to disinfectant agents and immune factors such as antibodies (27). Biofilm-producing bacteria could be the causative agents of an extensive variety of infections in animals and humans. Studies have reported 500 to 5,000 more resistance in biofilm-producing bacteria against antibiotic and disinfectant agents besides its planktonic forms (28). Through biofilm formation, bacteria can easily receive and disseminate resistance and virulence genes (29).
Several aquatic pathogens such as Vibrio spp., Flavobacterium spp., Aeromonas spp., and Yersinia spp. were detected as biofilm producers, as the surveillance of them in aquaculture environments was supported by this structure (30).
The complex process of biofilm formation in Aeromonas is a response to the specific environmental factors. Understanding the biofilm synthesis process by Aeromonas spp. could be useful to prevent the eradication of fish disease (31) and important to public health. The entrance of resistant and biofilm-producing bacteria by marine food consumption is a causative agent of chronic infection in humans. In the present study, the biofilm-formation ability of 19 strains of pathogenic A. hydrophila was evaluated by microtiter plate crystal violet in TSB as the best medium for bacterial biofilm production. Ormanci and Yucel demonstrated that different growth media, due to their diverse ingredients, had different effects on biofilm formation, with more favoring biofilm formation in less-rich growth media such as TSB (32). In another study, Chenia and Duma reported that the nutrient-rich condition could encourage strong biofilm production in A. culicicola isolates, but it was not for A. allosaccharophila isolates (31).
The establishment of suboptimal environmental conditions (improper temperature, nourishing, pH) for A. hydrophila could induce structural changes in bacterial surfaces and decrease attachment ability, viability, and pathogenicity depending on the isolation origin (8, 33). Apparently, the feasible condition for biofilm production is different in various microorganisms and the identification of these conditions and the inhibition of its establishment are important in any aquatic training system. In the present study, 100% (20/20) of the isolates were detected as biofilm-producer, which is a significant percentage. The data from the current study indicated that 79% of the Aeromonas isolates were moderate biofilm-producers within 96 h (0.088 < ODs < 0.176). Biofilm formation, a characteristic that can influence the pathogenicity of Aeromonas species, was detected in 71.4% of A. hydrophila isolates according to Guerra et al. (34). Another study by Igbinosa et al. showed that 86% of the isolates were biofilm-producers in the microtiter plate method (35). A similar conclusion was drawn by Ormanci and Yucel that reported biofilm formation by diverse Aeromonas species (food and environmental isolates) during 24 h (32). A study by Kirov et al., conducting in-vitro biofilm assays to assess biofilm formation on human tissue, reported that the ability of Aeromonas species to form biofilm further revealed the degree of their pathogenicity (36).
Antibiotic-resistant bacteria may cause some difficulties in the treatment process and help with the development of chronic infections. The indiscriminate use of antibiotics could be ending to the increase of resistant strains. Different types of antibiotics, particularly β-lactams, chloramphenicol, and tetracycline, are used to treat Aeromonas infections and an increase in resistance to these antibiotics is reported among pathogenic Aeromonas species (37). In the present study, all the isolates were susceptible to trimethoprim-sulfamethoxazole and ciprofloxacin. Several studies of antibiotic resistance have been performed in Aeromonas spp. isolated from food, clinical samples, untreated water, fish gut, and freshwater fish (38). Matyar et al. studied antimicrobial resistance in Aeromonas spp. isolated from water in Turkey and reported that 14.4% of the studied strains were resistant to tetracycline, 11.3% to chloramphenicol, and 7.2% to gentamycin (39). In contrast to the current study results, Dias et al. reported that all Aeromonas strains isolated from five animal species (red squirrel, snake, red deer, short-toed snake eagle, and tawny owl) were resistant to ciprofloxacin. This variation can be due to differences in host species and Aeromonas strains (40).
The present study investigated the correlation between antibiotic resistance and biofilm formation ability in the isolates using the Fisher test. There was no significant relationship between the biofilm-production rate and antibiotic resistance pattern because all the studied isolates were biofilm-producers, but a significant correlation was detected between the level of biofilm production and antibiotic resistance because all the strong biofilm-producing strains were resistant to 80% of the used antibiotics. In fact, the strong production of biofilm could protect bacteria against more antibiotics, possibly due to the protective structure or easy gene transfer in this structure. In the present study, the most resistance was detected to clindamycin and vancomycin in all producing strains. Clindamycin (such as macrolide and lincosamide) resistance is mainly due to one of these three mechanisms including the target site modification, ribosomal methylation, and mutation, which prevent the binding of antibiotics to its ribosomal target. Vancomycin resistance is caused by reduced vancomycin binding and failure to prevent cell wall synthesis. Certainly, we need complementary studies to provide evidence (16).
5.1. Conclusions
Antibiotic resistance and biofilm formation ability of Aeromonas species isolated from infected fish with clinical symptoms were elucidated. The increasing resistance to antibacterial agents could be produced by biofilm formation in bacteria, leading to persistent infections subsequently. Biofilm production is not the only route of resistance to antibiotics in bacteria, as, in our study, moderate resistance and resistance to some antibiotics were detected in some strains with strong biofilm production ability whereas moderate biofilm-producing bacteria were susceptible to these antibiotics. Aeromonas spp. are constantly considered as opportunistic pathogens. Therefore, the observation of hygiene in abattoir environments and aquaculture to decrease multidrug-resistant cloning in the environment and humans is important and recommended. A deeper knowledge of pathogenic bacteria (parameters influencing the virulence mechanisms) can provide new information to refine the existing treatment strategies to defy biofilm infections and develop novel treatment strategies.