1. Background
Staphylococcus aureus is a Gram-positive bacterium that is responsible for various infectious diseases, including cutaneous infections, endocarditis, pneumonia, empyema, osteomyelitis, and also some diseases due to toxins such as toxic shock syndrome, scalded skin syndrome and food poisoning. Staphylococcus aureus is an opportunistic pathogen and is also one of the most frequent agents of nosocomial infections. The infections due to S. aureus happened because of predisposing factors such as prolonged hospitalization, immune suppression, invasive medical procedures and chronic diseases (1-3). The first beta-lactam (β-lactam) antibiotic penicillin G was used in humans as a chemotherapeutic agent in 1941 (4). However, first cases of S. aureus resistance were reported in 1942 (5). These isolates carried a plasmid gene, blaZ that encoded a beta-lactamase enzyme (penicillinase) (1, 6). In 1961 a new drug named methicillin (penicillinase-stable semisynthetic penicillin) was introduced but methicillin-resistant S. aureus (MRSA) isolates were reported in the same year (7). Methicillin-resistant S. aureus isolates are resistant to all penicillins and other β-lactam antibiotics.
The expression of the mecA gene is responsible for the resistance of MRSA isolates to all β-lactam antibiotics by the synthesis of a protein, called PBP2a which has decreased affinity to β-lactams. The mecA gene is located in mobile genetic elements with high diversity identified as staphylococcal cassette chromosome mec (SCCmec) (8). The first SCCmec type was identified in Japan (9). Currently, 11 types of SCCmec elements have been identified (I-XI). Studies have found the SCCmec types I-V are the most frequent in the world. The MRSA isolates with SCCmec types I, II, III usually are responsible for nosocomial infections and are related to HA-MRSA (hospital-associated MRSA). SCCmec types II and III are long and possess multidrug resistance to isolates that carry these elements. The MRSA isolates with SCCmec types IV and V are related to CA-MRSA (community-acquired MRSA) have been spreading in communities that are isolated from patients with no obvious relation to hospitals.
SCCmec types III and IV are the most predominant types in Asian countries such as Iran and other continents, respectively. In recent years, CA-MRSA strains with SCCmec types IV have been spreading in a community in some areas of the world that has been created challenges for control and treatment of MRSA strains-related infections (2, 10-12). Accordingly, SCCmec typing is one of the most applicant methods for typing of MRSA isolates. SCCmec types have important roles in bacterial pathogenesis, thus the identification of SCCmec types of MRSA isolates may aid us in further diagnosis, determining the origins of infections, designing sufficient infection control systems for preventing and effective treatment of MRSA strains-related infections (10-12).
2. Objectives
To the best of our knowledge, there is no study regarding SCCmec typing of MRSA strains in Kermanshah Province, West of Iran, thus we aimed to evaluate the prevalence of MRSA, antibiotic susceptibility pattern and SCCmec types of MRSA strains isolated from hospitalized patients in a general hospital in Kermanshah Province, West of Iran.
3. Methods
3.1. Identification of Staphylococcus aureus Isolates
In this descriptive cross-sectional study, 146 cases were isolated from hospitalized patients in Imam Reza Hospital in Kermanshah Province, West of Iran from July 2016 to March 2017. The samples were obtained from sputum, blood, urine, wound, etc. The samples were transferred to the Microbiology Laboratory and immediately inoculated in Blood agar and Mannitol Salt agar plates (Merck GmbH, Darmstadt, Germany) and incubated at 35°C for 24 - 48 hours. The phenotypic and genotypic identification of S. aureus isolates carried out by diagnostic assays such as Gram staining, coagulase test, DNase test and detection of femB gene by PCR method (13).
3.2. Antimicrobial Susceptibility Testing
The antimicrobial susceptibility pattern of isolates was evaluated using the disk diffusion method based on Clinical and Laboratory Standard Institute (CLSI) guidelines (14). We used antibiotic disks (Mast Diagnostics Group Ltd, Merseyside, UK), including clindamycin (2 µg), vancomycin (30 µg), erythromycin (30 µg), amikacin (30 µg), gentamicin (10 µg), ciprofloxacin (5 µg), chloramphenicol (30 µg), trimethoprim-sulfamethoxazole (1.25 + 3.75 µg) and linezolid (30 µg). To perform the disk diffusion method, a microbial suspension equal to 0.5 McFarland was prepared from 18-hour old culture of S. aureus isolates. Then, sterile swaps were prepared from a microbial suspension on a surface of the Muller Hinton agar plates (Merck GmbH, Darmstadt, Germany) and, after 15 minutes of inoculation, the above antibiotic disks were placed at least 2.5 centimeters apart on the surface of the plates. The plates were incubated at 35°C - 37°C for 24 hours. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were applied as quality control strains in each batch of susceptibility testing. Multiple drug resistance (MDR) was defined as resistance to at least one agent in three or more antibiotic classes (15).
3.3. Determination of Vancomycin Resistance by Minimum Inhibitory Concentration Testing
Vancomycin resistance was determined by minimum inhibitory concentration (MIC) testing using the agar dilution method according to CLSI guidelines (14). The strains S. aureus ATCC 29213 and Enterococcus faecalis ATCC 29212 were used as quality control strains. Bacterial isolates were classified into vancomycin-sensitive S. aureus (VSSA), vancomycin-intermediate S. aureus (VISA) and vancomycin-resistant S. aureus (VISA) according to the MIC ranges ≤ 2 µg/mL, 4 - 8 µg/mL and MIC ≥ 16 µg/mL, respectively (14).
3.4. Identification of MRSA by Cefoxitin Disk Diffusion Test
All the S. aureus isolates were subjected to cefoxitin disk diffusion testing using a cefoxitin disk (30 µg). Inhibition zones diameter of ≤ 21 mm and ≥ 22 mm were considered MRSA and methicillin susceptible Staphylococcus aureus (MSSA), respectively (14). The efficacy of this test for the detection of MRSA isolates was compared with the results of PCR method.
3.5. PCR Analysis of Genes
All isolates were tested for selected genes, including femB, mecA, SCCmec types such as I, II, III, IVa, IVb, IVc, IVd, and V using specific primers (Table 1) (16-19). For PCR analysis of S. aureus isolates, total DNA was extracted using a High Pure PCR Template Preparation Kit (Roche, Basel, Switzerland). PCR amplification was carried out with automated thermal cycler (Applied Biosystems, USA) in a total volume of 25 µL containing 1 µL of Taq DNA polymerase, 2× Master Mix, 0.5 µM forward primer, 0.5 µM reverse primer, 1.5 mM MgCl2, 2 µL DNA template, and 8.5 µL nuclease-free water. The used PCR conditions were: initial denaturation at 95°C for 6 minutes, denaturation at 50 seconds, annealing at different degrees for each gene (Table 1) for 45 seconds, extension at 70°C for 50 seconds for 35 cycles and a final extension at 70°C for 10 minutes. Moreover, S. aureus reference strains NCTC10442, NCTC N315, NCTC 85/2082, NCTC CA05, and JCSC3624 were used as positive controls for SCCmec elements. Gel electrophoresis of PCR products carried out in 1.5% agarose gel at 85V for 45 min and visualized on an ultraviolet (UV) transilluminator (BioRad, USA).
| Genes | Primer | Sequences (5’ to 3’) | Product Size, bp | Annealing Temperature, °C | References |
|---|---|---|---|---|---|
| mecA | F | GTGAAGATATACCAAGTGATT | 147 | 62 | (16) |
| R | ATGCGCTATAGATTGAAAGGAT | ||||
| femB | F | CGTGAGAATGATGGCTTTGA | 388 | 55 | (17) |
| R | TTAATACGCCCATCCATCGT | ||||
| SCCmecI | F | GCTTTAAAGAGTGTCGTTACAGG | 613 | 60 | (17) |
| R | GTTCTCTCATAGTATGACGTCC | ||||
| SCCmecII | F | GATTACTTCAGAACCAGGTCAT | 287 | 62 | (18) |
| R | TAAACTGTGTCACACGATCCAT | ||||
| SCCmecIII | F | CATTTGTGAAACACAGTACG | 243 | 61 | (19) |
| R | GTTATTGAGACTCCTAAAGC | ||||
| SCCmecIVa | F | GCCTTATTCGAAGAAACCG | 776 | 63 | (16) |
| R | CTACTCTTCTGAAAAGCGTCG | ||||
| SCCmecIVb | F | TCTGGAATTACTTCAGCTGC | 493 | 63 | (16) |
| R | AAACAATATTGCTCTCCCTC | ||||
| SCCmecIVc | F | ACAATATTTGTATTATCGGAGAGC | 200 | 62 | (16) |
| R | TTGGTATGAGGTATTGCTGG | ||||
| SCCmecIVd | F | CTCAAAATACGGACCCCAATACA | 881 | 63 | (16) |
| R | TGCTCCAGTAATTGCTAAAG | ||||
| SCCmecV | F | ACCTACAGCCATTGCATTATG | 1159 | 63 | (18) |
| R | TGTATACATTTCGCCACTAGCT |
3.6. Statistical Analysis
Statistical analysis was performed using SPSS software version 16 for descriptive statistics of microbiological and clinical data. Statistical significance of differences between findings was evaluated by chi-square (χ2) test. The P value of less than 0.05 was considered statistically significant.
4. Results
In this descriptive cross-sectional study, between July 2016 and March 2017, 146 cases were isolated from hospitalized patients in Imam Reza Hospital in Kermanshah Province, West of Iran. Of 146 isolates, 126 isolates were confirmed as S. aureus using phenotypic methods and PCR analysis of femB gene. The highest rate of S. aureus isolates was collected from patients aged more than 46 years. Of 126 isolates, 53 cases (42.6%) were isolated from males and 73 cases (57.4%) from females. Among 126 cases isolated from the different hospital wards, the highest frequency related to the infectious ward (28.6%) and ICU (15.9%). Results of isolated S. aureus based on clinical source showed that the highest and lowest rate of S. aureus were isolated from trachea (22.9%) and urine (9.6%), respectively (Table 2).
| Variables | Total (N = 126) | MRSA (N = 83) | MSSA (N = 43) | P Value |
|---|---|---|---|---|
| Sex | 0.120 | |||
| Male | 53 (42.6) | 39 (47) | 14 (32.6) | |
| Female | 73 (57.4) | 44 (53) | 29 (67.4) | |
| Age group, y | 0.015 | |||
| ≤ 15 | 13 (10.3) | 8 (9.6) | 5 (11.6) | |
| 16 - 30 | 18 (14.3) | 6 (7.2) | 12 (27.9) | |
| 31 - 45 | 15 (11.9) | 9 (10.8) | 6 (14) | |
| 46 - 60 | 50 (39.7) | 37 (44.6) | 13 (30.2) | |
| ≥ 61 | 30 (23.8) | 23 (27.7) | 7 (16.3) | |
| Wards | 0.995 | |||
| Infectious | 36 (28.6) | 25 (30.1) | 11 (25.6) | |
| ICU | 20 (15.9) | 14 (16.5) | 6 (14) | |
| Surgery | 16 (12.7) | 10 (12) | 6 (14) | |
| Children | 15 (11.9) | 9 (10.8) | 6 (14) | |
| Dialysis | 16 (12.7) | 9 (10.8) | 7 (16.3) | |
| Other wards | 11 (8.7) | 8 (9.6) | 3 (7) | |
| Source of samples | 0.116 | |||
| Tracheal | 25 (19.8) | 19 (22.9) | 6 (14) | |
| Pus | 23 (18.3) | 13 (15.7) | 10 (23.3) | |
| Blood | 18 (14.3) | 10 (12) | 8 (18.6) | |
| Urine | 17 (13.6) | 8 (9.6) | 9 (20.9) | |
| Surgical | 16 (12.7) | 11 (13.3) | 5 (11.6) | |
| CSF | 14 (11.1) | 13 (15.7) | 1 (2.3) | |
| Pleura | 13 (10.3) | 9 (10.8) | 4 (9.3) |
aValues are expressed as No. (%).
PCR amplification of mecA gene showed that of 126 S. aureus isolates, 83 cases (65.9%) were MRSA and 43 cases (34.1%) were MSSA. Eighty-one cases (64.3%) of isolates were MRSA using cefoxitin diffusion disk test which showed the sensitivity and specificity of this phenotypic test compared to results of PCR analysis of mecA gene as the gold standard were 97.6% and 100%, respectively. Findings of antimicrobial susceptibility testing of S. aureus isolates in this study showed that all isolates were sensitive to vancomycin by both methods: disk agar diffusion and MIC testing by agar dilution method (MIC ≤ 2 µg/mL). The highest resistance rate was related to erythromycin (75.4%) and ciprofloxacin (73%). There was a statistically significant difference in antibiotic susceptibility pattern of MRSA and MSSA isolates for some antibiotics such as gentamicin, amikacin, erythromycin, ciprofloxacin, and linezolid (P < 0.05). All MRSA isolates were MDR (Table 3).
| Antibiotics | Total (N = 126) | MRSA (N = 83) | MSSA (N = 43) | P Value |
|---|---|---|---|---|
| Gentamicin | 41 (32.5) | 35 (42.2) | 6 (13.9) | < 0.05 |
| Amikacin | 39 (30.9) | 33 (39.7) | 6 (13.9) | < 0.05 |
| Erythromycin | 95 (75.4) | 73 (87.9) | 22 (51.2) | < 0.05 |
| Ciprofloxacin | 92 (73) | 71 (85.5) | 21 (48.8) | < 0.05 |
| Clindamycin | 70 (55.6) | 52 (62.6) | 18 (41.8) | > 0.05 |
| Trimethoprim/sulfamethoxazole | 48 (38.1) | 36 (43.4) | 12 (27.9) | > 0.05 |
| Tetracycline | 44 (34.9) | 35 (42.2) | 9 (20.9) | > 0.05 |
| Linezolid | 4 (3.2) | 4 (100) | 0 | < 0.05 |
| Vancomycin | 0 | 0 | 0 |
aValues are expressed as No. (%).
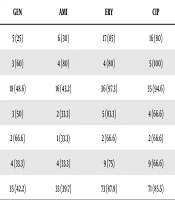
Of 83 MRSA isolates, SCCmec types were detected by PCR as follows: 20 cases (24.1%) type I, 5 cases (6%) type II, 37 cases (44.6%) type III (the most prevalent type), 6 cases (7.2%) type IVa, and 3 cases (3.6%) type IVc. The SCCmec types IVb, IVd, and V were detected in no isolate. Also, 12 cases (14.5%) of isolates could not be typed by this method. The prevalence of HA-MRSA (types I, II, and III) and CA-MRSA (types IV and V) was 74.7% and 10.8%, respectively. Most SCCmecIII isolates were resistant to erythromycin, ciprofloxacin, clindamycin, and tetracycline (Table 4).
| Sccmec Type | Cases | GEN | AMI | ERY | CIP | CLI | TRIM/SU | TET | LIN |
|---|---|---|---|---|---|---|---|---|---|
| I | 20 (24.1) | 5 (25) | 6 (30) | 17 (85) | 16 (80) | 8 (40) | 10 (50) | 6 (30) | 1 (5) |
| II | 5 (6) | 3 (60) | 4 (80) | 4 (80) | 5 (100) | 3 (60) | 2 (40) | 2 (40) | - |
| III | 37 (44.6) | 18 (48.6) | 16 (43.2) | 36 (97.3) | 35 (94.6) | 31 (83.8) | 19 (51.3) | 25 (67.5) | 2 (5.4) |
| IVa | 6 (7.2) | 3 (50) | 2 (33.3) | 5 (83.3) | 4 (66.6) | 2 (33.3) | 2 (33.3) | - | - |
| IVc | 3 (3.6) | 2 (66.6) | 1 (33.3) | 2 (66.6) | 2 (66.6) | 2 (66.6) | - | - | - |
| Nontypable | 12 (14.5) | 4 (33.3) | 4 (33.3) | 9 (75) | 9 (66.6) | 6 (50) | 3 (25) | 2 (16.6) | 1 (8.3) |
| MRSA total | 83 (100) | 35 (42.2) | 33 (39.7) | 73 (87.9) | 71 (85.5) | 52 (62.6) | 36 (43.4) | 35 (42.2) | 4 (4.8) |
Abbreviations: AMI, amikacin; CIP, ciprofloxacin; CLI, clindamycin; ERY, erythromycin; GEN, gentamicin; LIN, linezolid; MRSA, methicillin-resistant Staphylococcus aureus; TET, tetracycline; TRIM/SU, trimethoprim sulfamethoxazole.
aValues are expressed as No. (%).
5. Discussion
Methicillin-resistant S. aureus is an important agent of various infectious diseases, especially bacterial nosocomial infections. In recent years, increasing prevalence of MRSA isolates has created many problems for the treatment of infections caused by this bacterium (1). The mecA gene is responsible for the resistance to all β-lactam antibiotics located in a mobile genetic element identified as SCCmec. SCCmec typing has been developed for the typing of MRSA isolates (3, 10-12). In this research, we investigated the prevalence of MRSA, the antibiotic susceptibility pattern, and the prevalence of SCCmec types in MRSA strains isolated from hospitalized patients in a general hospital in Kermanshah Province, West of Iran. In this research, we investigated the association of demographic data with MRSA isolates. However, most of these variables were not found to have a significant association with MRSA isolates. We found only a significant association between age groups and frequency of MRSA. The results of this research revealed the highest rate of MRSA isolates was collected from patients aged more than 46 years. It could be due to decreasing resistance and increasing exposure to healthcare systems in the elderly (20).
Findings of the antibiotic susceptibility testing in this research revealed that all S. aureus isolates were sensitive to vancomycin by both disk agar diffusion and MIC testing. The resistance rate to linezolid was 3.2%. These results showed vancomycin and linezolid were still effective drugs for the treatment of MRSA associated infections. Results of the prevalence of MRSA isolates using identification of mecA gene and cefoxitin disk diffusion test were 65.9% and 64.3%, respectively. The sensitivity and specificity of cefoxitin disk diffusion test compared to the results of PCR analysis of mecA gene as the gold standard were 97.6% and 100%, respectively. This finding is similar to previous studies, which showed the suitability of cefoxitin disk diffusion test for detection of MRSA isolates (21, 22). Broekema et al. (21) reported the sensitivity and specificity of cefoxitin disc diffusion test 97.3% and 100%, respectively and Velasco et al. (22) reported the sensitivity and specificity of this test 100% compared to mecA PCR method as the gold standard for detection of MRSA isolates. In this regard, CLSI has recently recommended cefoxitin disc diffusion test for phenotypic detection of MRSA isolates (14). These results showed this phenotypic method could be considered a simple, cheap and reliable test for the identification of MRSA isolates in all laboratories.
Methicillin-resistant S. aureus prevalence is high (65.9%) in this study and higher than other studies were done in different regions in Iran (23). The highest rates of MRSA are found in Asia, North and South America (> 50%). Intermediate rates (25% - 50%) are seen in China, Australia, Africa, and some European countries and low rates of MRSA are reported from Netherlands and Scandinavia. The differences in the frequency of the MRSA isolates could be due to the diversity of studied populations, clinical specimens and also different infection control policies (2). Comparing to the results of antibiotic susceptibility testing between MRSA and MSSA isolates showed that all MRSA isolates were MDR. There was a statistically significant difference in antibiotic susceptibility pattern of MRSA and MSSA isolates for some antibiotics such as gentamicin, amikacin, erythromycin, ciprofloxacin, and linezolid (P < 0.05). These results are similar to the findings of the previous study were carried out in Ethiopia and India (24-26).
The prevalence of HA-MRSA (types I, II, and III) in this study was 74.7%. Our findings regarding SCCmec typing by PCR method showed that SCCmec III was the most prevalent type in this study (44.6%). These results were consistent with previous studies regarding the predominance of SCCmec type III in other regions in Iran (27, 28), and the other Asian countries (12). The next predominant type in this study was SCCmec type I (24.1%). The prevalence rate of SCCmec type I in this study was higher than other studies performed in Iran (28). SCCmec type I is the first identified type that was reported from a few countries, including Iran, Japan, and Brazil (28-30). These SCCmec types are associated with HA-MRSA, regarding our isolates were obtained from clinical specimens of hospitalized patients and likely most of these isolates have originated from a unique ancestor. According to the findings of the current study, most of SCCmec III harboring MRSA isolates were resistant against selected antibiotics such as tetracycline and erythromycin. The SCCmec type III is a long mobile genetic element and carries many antibiotic-resistance genes and determinants such as transposon Tn554 and plasmid pT181 that encode tetracycline and erythromycin resistance (12).
5.1. Conclusions
In general, the results of this study showed that the prevalence of MRSA isolates in Kermanshah Province, West of Iran, is high and the cefoxitin diffusion disk testing could be considered a simple, cheap and reliable test for identification of MRSA isolates in all laboratories. The resistance rate to most antibiotics in MRSA isolates was higher than MSSA isolates. All MRSA isolates were recognized as MDR. Thus, the therapeutic effects of all available drugs must be applied for the treatment of infections caused by MRSA isolates. The most frequent types of SCCmec in this study were type III and I. These SCCmec types are associated with HA-MRSA; regarding our isolates were obtained from clinical specimens of hospitalized patients. The continuous surveillance of antibiotic resistance patterns of S. aureus strains, especially MRSA isolates should be considered an important necessity. The limitation of our study was the impossibility of using other typing methods such as pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) for MSSA and MRSA isolates.