1. Background
Acinetobacter is classified among gram-negative, immobile coccobacilli, which is encapsulated, forced aerobic, spore-free, and unable to ferment glucose. This bacterium is commonly found in soil and water (1, 2). It is an opportunistic pathogen that does not cause disease in healthy individuals, while it is easily transmitted through the population, urging hospitals to improve their methods of infection control. As a nosocomial pathogen, Acinetobacter baumannii mainly affects the patients admitted to the intensive care unit (ICU), such as patients with trauma, injuries, and burns and those requiring mechanical ventilation (3).
Resistance mechanisms may vary in different species of Acinetobacter. One of these mechanisms is the production of beta-lactamase enzymes, which are inactivated by the hydrolysis of the central nucleus in beta-lactam antibiotics. There are several types of beta-lactamases, including sulfhydryl variable, pseudomonas extended resistant, cefotaximase, temoneira, and AmpC β-lactamase. In general, the genes encoding these enzymes are located on the plasmid. Other classes of beta-lactamases have emerged with the increasing use of antibiotics (e.g., cephalosporins), which are known as extended-spectrum β-lactamases (ESBLs) and have a wider spectrum of activity compared to primary beta-lactamases. These enzymes could hydrolyze antibiotics such as penicillin, cephalosporins, and aztreonam (4, 5).
Acinetobacter also contain carbapenem-hydrolyzing oxacillinase enzymes, which make bacteria resistant to carbapenems and penicillins (6). In 2011, a report suggested that various genes encoding resistance against different drugs were identified in Acinetobacter, including beta-lactams, tetracyclines, and glycopeptides (7, 8). Acinetobacter species demonstrated different behaviors in terms of antibiotic resistance. In addition, the inherent resistance of the bacterium to numerous common antibiotics helps this opportunistic pathogen quickly acquire other resistance mechanisms in response to broad-spectrum antibiotics.
Antibiotic resistance assessment provides physicians with beneficial data for the selection of effective experimental treatments. Therefore, it is essential to identify resistant bacterial strains in a specific geographical region to prevent their spread.
2. Objectives
The present study aimed to investigate the antibiotic resistance pattern of the isolated Acinetobacter strains from the patients admitted to various wards of Imam Reza hospital in Kermanshah, Iran.
3. Methods
In this descriptive, cross-sectional study, a laboratory expert provided the list of the patients admitted to Imam Reza hospital in Kermanshah, Iran during 2016-2018 who had positive culture samples of Acinetobacter (sputum, urine, blood, wound, fecal, stomach, and eye samples). The susceptibility pattern and antibiotic resistance of the pseudomonas isolated from each patient were reported, and the samples were sent to the microbiology laboratory, grown in eosin methylene blue and blood agar, and incubated at the temperature of 37 °C for 24 hours. After colony growth and the initial identification of the organism, the bacterium was identified based on the colony color, presence/absence of hemolysis, and gram-smear staining prepared from the colonies.
At the next stage, antibiogram was performed using the disc-diffusion method, and the exact diameter of the growth inhibition zone was determined in millimeters using the Müller-Hinton agar medium. In addition, comparison was performed with the Clinical and Laboratory Standard Institute (CLSI) standard table for gentamicin (10 µg), ciprofloxacin (5 µg), ceftazidime (30 µg), cotrimoxazole (1.25 µg), ceftriaxone (30 µg), cefepime, imipenem, ampicillin (10 µg), ampicillin-sulbactam (10 µg), amikacin (30 µg), cefixime (5 µg), and tazobactam (110 µg).
The bacterial suspension was prepared with turbidity equivalent to 0.5 McFarland of the tube turbidity (× 108 CFU/mL 1.5), and the lawn culture was performed in triplicate using a sterile swab on a plate containing the Müller-Hinton medium. In addition, pliers were utilized to remove the discs from the freezer one hour before they were placed on the culture medium and stabilized with the tip of a pair of pliers; afterwards, the plates were incubated at the temperature of 37 °C for 24 hours. To read the results, the diameter of the inhibition zone was measured in millimeters using an accurate ruler, and the results were reported as sensitivity (S1), resistance (R2), and semi-sensitive zones (I3).
In this research, all cases of ethics in research and ethical requirements have been respected and the confidentiality of information has been preserved.
3.1. Statistical Analysis
Data analysis was performed in SPSS version 18 using descriptive statistics (mean, standard deviation, frequency, and percentage). In addition, chi-square (χ2) and independent t-test were applied to evaluate the correlations between antibiotic resistance, infection prevalence, and infection severity. In all the statistical analyses, P-value of 0.05 was considered significant.
4. Results
In 2016, 2017, and 2018, a total of 231, 271, and 224 positive cases of Acinetobacter were reported in the patients admitted to Imam Reza hospital, respectively. Out of 726 patients with Acinetobacter positive cultures during these periods, 58.1% were male, and 47.1% were female with the mean age of 60.57 ± 20.99 years.
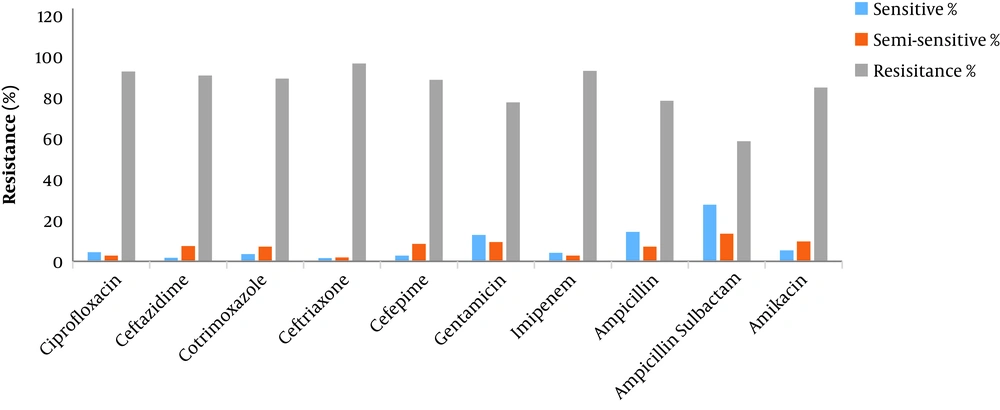
Among the positive culture samples, 73.3% were sputum samples, 10.1% were urine samples, 6.6% were blood samples, 5.1% were wound samples, 4.7% were fecal samples, and 0.1% were stomach and eye samples. Table 1 shows the number of the positive Acinetobacter cases by hospital sections. The antibiogram results are also shown in Table 2 and Figure 1. According to the findings, the organism was resistant to all the antibiotics, with the highest sensitivity rate observed against ampicillin-sulbactam (27.7%).
| Hospital Units | Percentage (%) |
|---|---|
| ICUs (general, neonatal, pediatric) | 75.88 |
| Emergency department | 7.16 |
| Internal (internal, infections, pediatric, neurology) | 11.55 |
| Surgery (general, gynecological, urology) | 4.26 |
| Outpatients | 0.68 |
| Transplantation (bone marrow, kidney) | 0.41 |
| Total | 100 |
| Antibiotic | Sensitive (%) | Semi-sensitive (%) | Resistant (%) | No. (%) of Tested Samples on Antibiotics |
|---|---|---|---|---|
| Ciprofloxacin | 4.5 | 2.8 | 92.7 | 536 (73.8) |
| Ceftazidime | 1.8 | 7.4 | 90.8 | 542 (74.7) |
| Cotrimoxazole | 3.6 | 7.1 | 89.3 | 169 (23.3) |
| Ceftriaxone | 1.6 | 1.9 | 96.6 | 640 (88.2) |
| Cefepime | 2.8 | 8.5 | 88.7 | 248 (34.2) |
| Gentamicin | 12.9 | 9.4 | 77.7 | 556 (76.6) |
| Imipenem | 4.1 | 2.8 | 93 | 603 (83.1) |
| Ampicillin | 14.4 | 7.2 | 78.4 | 125 (17.2) |
| Ampicillin-sulbactam | 27.7 | 13.5 | 58.7 | 155 (21.3) |
| Amikacin | 5.4 | 9.7 | 84.9 | 186 (25.6) |
5. Discussion
The multiple-drug resistance of A. baumannii has led to numerous medical issues in patient treatment. The bacterial infection is associated with variable susceptibility to different antibiotics and is influenced by environmental causes and complex patterns of antibacterial use. The development and spread of resistant bacterial species in different hospital wards may have been caused by various patterns of antibiotic use and lack of sufficient resources to control hospital infections (9, 10).
In the present study, most of the samples were isolated from sputum and trachea, indicating that the respiratory tract is most commonly affected by the infections caused by Acinetobacter. Therefore, the infection could be controlled and prevented by the disinfection and sterilization of respiratory equipment and instruments (e.g., respiratory catheters). Our findings also indicated that the Acinetobacter strains that were isolated from hospitalized patients were resistant to the antibiotics that are widely used. Furthermore, some of the bacterial strains were immune to many antibiotics tested simultaneously, and the treatment of the infections caused by these organisms is extremely challenging.
The infections caused by A. baumannii have become more complex with the increased resistance of infection-causing strains to several antibiotics. Due to the excessive use of third-generation cephalosporins and no observance of hygienic principles in the community, which were also highlighted by our findings, considerable resistance has been reported against third-generation cephalosporins. Given the estimated 96.6% resistance to ceftriaxone and 90.8% resistance to ceftazidime, it could be inferred that third-generation cephalosporins are not effective in the treatment of the infections caused by A. baumannii.
In the current research, elevated carbapenem resistance (imipenem) demonstrated the indiscriminate use of these drugs regardless of the risks associated with medication resistance. In recent decades, carbapenem-resistant bacterial strains have been a monumental challenge in the ICU treatment of Acinetobacter infections. In this regard, Dent et al. reported that resistance of Acinetobacter strains to imipenem was observed in 29% and 41% of the isolates, which were resistant to aminoglycosides, cephalosporins, broad-spectrum penicillins, and quinolones (11).
In the current research, the rate of antibiotic resistance was higher due to the extensive use of imipenem in multiple hospital wards, as well as the indiscriminate and inappropriate drug usage by the population. On the other hand, the quality of antibiotic discs and the applied techniques to determine susceptibility in microbiology laboratories may lead to differences in the obtained results in various geographical areas, even within the same country (12). In 2002 - 2004, another study investigated the resistance of Acinetobacter isolated from Iraqi and Afghan soldiers, and 65% of the isolates were reported to be immune to imipenem, while the prevalence rate was lower compared to our findings (13). Another study in this regard indicated that among 52 bacterial strains, all the isolates were resistant to piperacillin, piperacillin-tazobactam, ticarcillin, clavulanic acid, cefepime, cefotaxime, ceftazidime, ceftriaxone, gentamicin, and ciprofloxacin. In the mentioned study, resistance to ciprofloxacin, ampicillin-sulbactam, cotrimoxazole, and amikacin was also observed in 8, 55, 66, and 74% of the strains, respectively (14).
In infection control, patients should be colonized with multidrug-resistant Acinetobacter strains. In Asia and the Mediterranean region, numerous cases of nosocomial drug-resistant A.baumannii strains have been identified, and various forms of carbapenemases have also been reported in these countries (15). The isolates studied in the latest research in this regard demonstrated an extremely high rate of imipenem resistance, which seems to be increasing progressively as many of the treatment schemes for nosocomial infections encompass imipenem. Only if the organism is resistant to other antibiotics could the antibiotic be used as the 'last resort'. On the other hand, a new type of metallo beta lactamase known as New Delhi metallo-beta-lactamase 1 (NDM-1) is known to many members of the Enterobacteriaceae and A. baumannii families. NDM-1-producing bacteria are resistant to multiple groups of antibiotics, including fluoroquinolones, aminoglycosides, and beta-lactams (especially carbapenems), while they are only sensitive to colistin and tigecycline (16).
The CDC has proposed recommendations for the prevention of multidrug-resistant pathogens in ICUs; some of the suggested measures are hand washing, treatment in patient interaction before they show a negative culture for the target pathogens, active and regular surveillance cultures, training of healthcare providers, and safe cleaning of the hospital environment polluted by these pathogens (17-19).
5.1. Conclusion
According to the evaluation of antimicrobial resistance, all the bacterial isolates had multiple-drug resistance and were resistant to ciprofloxacin, ceftazidime, cotrimoxazole, ceftriaxone, cefepime, gentamicin, imipenem, ampicillin, ampicillin-sulbactam, and amikacin. Due to the extremely rapid growth of antibiotic resistance due to the indiscriminate use of these medications (especially in developed countries), it is essential to assess the resistance pattern of pathogenic species (particularly in patients with nosocomial infections) and identify multidrug-resistant species. Moreover, determining the antibiotic resistance patterns that are specific to each geographical area could be beneficial in the planning of treatment protocols to prevent the emergence and spread of multidrug-resistant organisms.