1. Background
Pneumonia caused by the new coronavirus was announced as the coronavirus disease 2019 (COVID-19) by the World Health Organization (WHO) on February 11, 2020, which was taken to the epidemic scale and spread rapidly all over the world (1). Initially, the exact diagnosis of COVID-19 was possible by detecting the SARS-CoV-2 ribonucleic acid (RNA) in the specimens sent from the infected cases (2). Later, various diagnostic tests were used to identify the virus, including molecular tests such as reverse transcription-polymerase chain reaction (RT-PCR) tests, which identifies the genetic material of the virus, and antigen tests that detect special proteins on the virus surface. Today, new tests have been introduced for COVID-19 detection, which provide diagnostic benefits (3).
In order to adequately cope with the COVID-19 pandemic and remove the constraints imposed on the community, there is an urgent need for tests that can work with a minimum of devices and equipment, including at home. COVID-19 IgG/IgM antibody tests are used for the detection of SARS-CoV-2 with quick point-of-care (POC) tests. POC diagnostic test methods are rapid and use a mucus sample from the nose or throat. As such, POC tests that are scalable, cost-effective, and easy to use are critical to battle COVID-19 (4).
Optimized enzyme-linked immunosorbent assay (ELISA) is used in numerous clinical virology laboratories for diagnostic purposes and evaluating conditions such as infections, vaccination, and re-infection (5, 6). Most of the antibodies reported for SARS-CoV-2 have been used for ELISA (7). Furthermore, serological tests for the detection of SARS-CoV-2 are under development. Zhang et al. determined IgG and IgM positivity from COVID-19 patient sera using the N protein-specific ELISA test (8).
In the study conducted by Huang et al., the results of the unenhanced computed tomography (CT) of 41 patients with definite COVID-19 were published as the first article in this regard (9). Following that, scientific publications on the diagnosis of COVID-19 increased, and the clinical indications for chest CT are constantly changing. Although RT-PCR is the ‘gold standard’ diagnostic test for confirming the diagnosis of COVID-19, it could be a valuable guide in the initial assessment of non-contrast chest CT patient populations (10).
Accurate detection of the global epidemic-causing COVID-19 is imperative for disease surveillance and control. Dozens of tests have been made available within a short time for COVID-19 diagnosis. With the rapid development of such tests, rigorous evaluations have become difficult, and there are important uncertainties regarding the specificity of these tests. Quantitative RT-qPCR has been accepted as a reference test and is widely used for COVID-19 diagnosis. However, RT-qPCR applications are not possible in all health centers. Therefore, tests with widespread use in the diagnosis of infectious disease should be evaluated from all perspectives to assess their potential role in the prognosis and management of COVID-19 patients (2). Our study is of particular importance as it clarifies the mentioned situation.
2. Objectives
This meta-analysis aimed to compare the diagnostic sensitivity of POC, ELISA, and CT tests in the diagnosis of COVID-19 infection, evaluate the quality of the existing tests, and compare the PCR-verified sensitivity of these test methods.
3. Methods
3.1. Protocol and Registration
The preferred reporting items for systematic reviews and meta-analysis (PRISMA) protocol was used for this review. The focus of the review was the comparison of the sensitivity of the diagnostic methods used for COVID-19.
3.2. Search Strategy
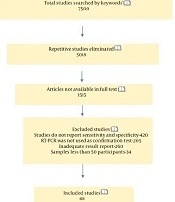
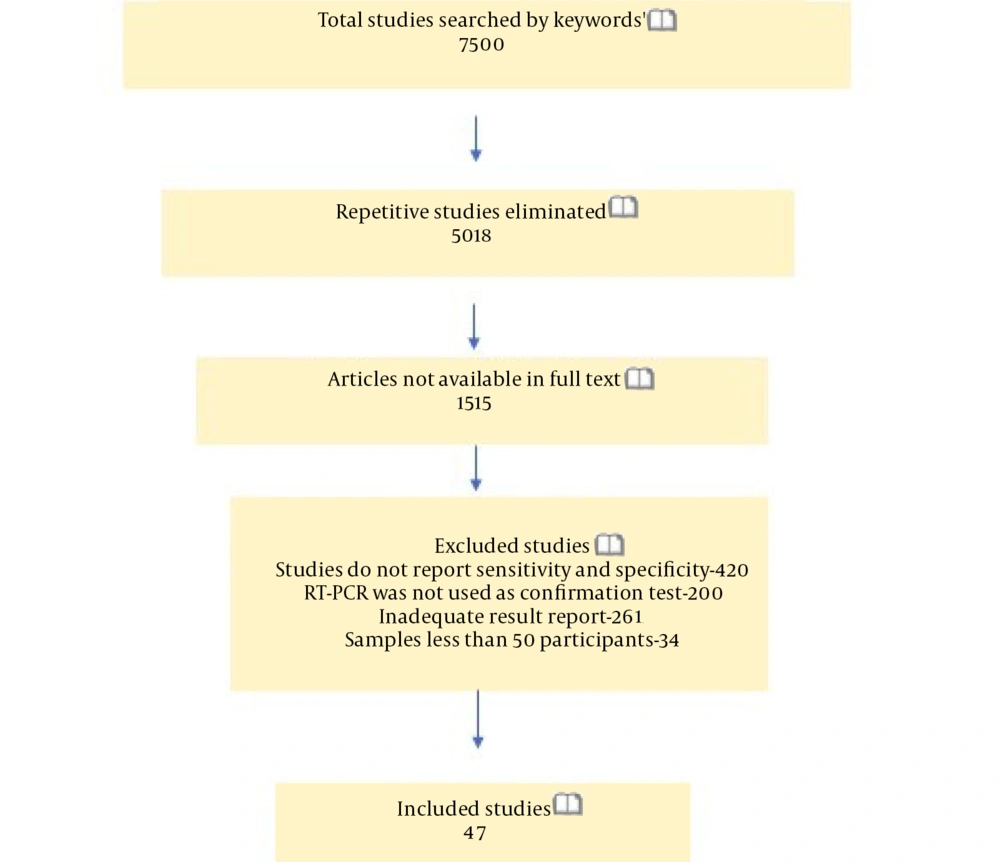
This meta-analysis was conducted via searching in databases such as NCBI, Google Scholar, and Medline for studies published in English in 2020. Subject headings/sub-headings were used for the COVID-19/SARS-CoV-2 and the POC, ELISA, and CT tests. The literature review applied to publications during January 1, 2020-November 15, 2020. We also evaluated the articles included in the references of other studies (Figure 1).
3.3. Eligibility Criteria
The eligibility criteria of our study were clinical researches reporting the sensitivity of POC, ELISA, and CT in COVID-19 diagnosis. Review studies, letters to the editor, case reports, modeling/economic studies, articles with a smaller sample size than 50, studies lacking clear data, and those not comparing the mentioned test methods with PCR were excluded.
3.4. Study Selection
Articles were searched using keywords in the literature search via Google Scholar, NCBI, and Medline. At this stage, we collected the articles identified in these databases, and the articles that did not meet the inclusion criteria were excluded. Following that, the full texts of the eligible articles were retrieved and reviewed independently. Disagreements between the authors were resolved by scientific consensus. In this meta-analysis, we selected the studies that compared the sensitivity rate of the test methods used for COVID-19 diagnosis with a reference RT-PCR standard. Clinical trials conducted on 50 or more patients reporting the sensitivity of POC, ELISA, and CT for COVID-19 diagnosis were selected and reviewed.
3.5. Quality Assessment
Table 1 shows the methodological evaluation of the study. Notably, these quality criteria were not considered as exclusion criterion. The qualitative review of the selected studies for the meta-analysis was scored based on a checklist designed with critical evaluation by three independent researchers. These criteria were the applied test method, number of the included patients, and the false negative and true positive rates of the applied tests (Table 1).
| 3 Points | 2 Points | 1 Point | |
|---|---|---|---|
| Test method | ELISA | CT | POC |
| No. of patients | > 500 | 100 - 500 | < 100 |
| False-negative rate of used test, % | < 10 | 10 - 30 | > 30 |
| True-positive rate of used test, % | > 80 | 50 - 80 | < 50 |
3.6. Data Extraction
One of the researchers extracted the aggregated data using a standard electronic data entry form. For each study, the second and third investigators validated the entered data. Duplicate data were not found, and data were collected on trace characteristics (country, sample type, applied method) and the methodological details regarding the index and reference.
3.7. Statistical Analysis
Data analysis was performed in SPSS version 25 and comprehensive meta-analysis (CMA) using one-way ANOVA and paired sample t-test, which were performed in SPSS. The effect sizes and heterogeneity (I2 and Q) of the selected studies were calculated in CMA, and a forest plot was used to assess publication bias. Heterogeneity in meta-analysis refers to variations in the results of the selected studies. The thresholds for the interpretation of I2 could be misleading as the significance of the discrepancy depends on several factors. A rough guide to such interpretation is as follows:
• 0 - 40%: may not matter;
• 30 - 60%: may represent moderate heterogeneity;
• 50 - 90%: may represent substantial heterogeneity;
• 75 - 100%: significant heterogeneity
3.8. Data Report
In the first part of our analysis, we estimated the sensitivity of POC, ELISA, and CT test methods and categorized the studies based on their applied tests methods. In several studies, researchers evaluated the accuracy of multiple test methods simultaneously (eg, POC and ELISA). For the index test performed on each study, the required numbers to form 2 × 2 probability tables were extracted. Each assessment of a particular index test was considered as the arm of the study. For instance, a study performing two ELISA and three CT evaluations on the same patient group would contribute to five study arms. It was also assumed that the sensitivity results of the tests would vary depending on the methods, countries, and sample types. As such, we classified the samples with primary results based on the applied methods, regions, and countries.
4. Results
4.1. Study Selection
The literature search yielded 7,500 studies corresponding to the keywords. After eliminating repetitive studies, 5,018 articles remained (Figure 1). In total, 47 studies that met the inclusion criteria were included, and review studies, letters to the editor, case reports, modeling/economic studies, articles without clear data, the studies with smaller sample sizes than 50, and those without a PCR comparison were excluded. Most of the studies that were selected for the meta-analysis (n = 47) and evaluated POC, ELISA, and CT methods were conducted in China. In the studies selected for the meta-analysis, CT was the most commonly applied method (Table 2).
| Study | Country | COVID-19 Diagnosis Method | Sample | Number of Tests (N) | Qualitative Score | ||
|---|---|---|---|---|---|---|---|
| Total | Positive with PCR | Positive with CT, ELISA and POC | |||||
| Ai et al. (11) | China | CT | Throat swab | 1014 | 601 | 580 | 11 |
| Adams et al. (12) | ABD | ELISA | Blood | 142 | 40 | 34 | 10 |
| Cassaniti et al. (13) | Italy | POC | Blood | 50 | 41 | 7 | 4 |
| Virgilio Paradiso et al. (14) | Italy | POC | Blood | 191 | 70 | 33 | 5 |
| Perez-Garcia et al. (15) | Spain | POC | Blood | 90 | 55 | 26 | 4 |
| Liu et al. (16) | China | POC | Pharyngeal swab | 179 | 90 | 77 | 8 |
| Zhao et al. (17) | China | ELISA | Blood | 173 | 173 | 161 | 11 |
| Lou et al. (18) | China | ELISA | Blood | 80 | 80 | 79 | 10 |
| Guo et al. (19) | China | ELISA | Blood | 82 | 82 | 65 | 8 |
| Infantino et al. (20) | Italy | ELISA | Nasopharyngeal swab | 64 | 64 | 41 | 7 |
| Jin et al. (21) | China | ELISA | Sputum/oral swab | 76 | 27 | 24 | 9 |
| Cai et al. (22) | China | ELISA | Oral swab | 276 | 276 | 225 | 10 |
| Xie et al. (23) | China | ELISA | Blood | 56 | 16 | 15 | 10 |
| Yangchun et al. (24) | China | ELISA | Blood | 284 | 205 | 197 | 11 |
| Qian et al. (25) | China | ELISA | Blood | 972 | 503 | 486 | 12 |
| Fang et al. (26) | China | CT | Throat swab | 81 | 51 | 50 | 9 |
| Ruch et al. (27) | Fransa | CT | Unspecified | 572 | 572 | 558 | 11 |
| Xiang et al. (28) | China | CT | Unspecified | 83 | 53 | 50 | 8 |
| Wang et al. (29) | China | CT | Unspecified | 107 | 90 | 50 | 7 |
| Du et al. (30) | China | CT | Unspecified | 125 | 125 | 99 | 9 |
| Rousan et al. (31) | Jordan | CT | Nasopharyngeal swab | 88 | 88 | 75 | 7 |
| Shabrawishi et al. (32) | Saudi Arabia | CT | Unspecified | 150 | 150 | 119 | 8 |
| Werberich et al. (33) | Brasil | CT | Unspecified | 78 | 78 | 48 | 6 |
| Bernheim et al. (34) | China | CT | Unspecified | 121 | 121 | 94 | 8 |
| Wu et al. (35) | China | CT | Unspecified | 80 | 80 | 73 | 8 |
| Xu et al. (36) | China | CT | Unspecified | 90 | 90 | 69 | 8 |
| Cui et al. (37) | China | CT | Unspecified | 95 | 95 | 53 | 7 |
| Falaschi et al. (38) | Italy | CT | Unspecified | 773 | 462 | 419 | 11 |
| Imai et al. (39) | Japan | POC | Blood | 112 | 112 | 37 | 5 |
| Nie et al. (40) | China | CT | Unspecified | 163 | 163 | 145 | 7 |
| Parry et al. (41) | India | CT | Unspecified | 147 | 147 | 51 | 7 |
| Goldberg-Stein et al. (42) | ABD | CT | Nazal swab | 141 | 141 | 80 | 7 |
| Gu et al. (43) | China | CT | Nasopharyngeal, oropharyngeal swab | 50 | 50 | 38 | 8 |
| Meiler et al. (44) | Germany | CT | Unspecified | 64 | 64 | 62 | 7 |
| Brendish et al. (45) | UAE | POC | Nasal swab | 517 | 499 | 157 | 6 |
| Imai et al. (39) | Japan | CT | Blood | 112 | 112 | 77 | 8 |
| Butt et al. (46) | Pakistan | POC | Nasopharyngeal swab | 70 | 45 | 43 | 8 |
| Haq et al. (47) | India | POC | Nasopharyngeal swab | 84 | 72 | 62 | 7 |
| Mohon et al. (48) | Canada | POC | Nasopharyngeal swab | 120 | 42 | 41 | 9 |
| Ghofrani et al. (49) | ABD | POC | Nasal swab/nasopharyngeal swab | 112 | 17 | 16 | 9 |
| Smithgall et al. (50) | ABD | POC | Nasal swab/nasopharyngeal swab | 113 | 113 | 84 | 7 |
| Bulterys et al. (51) | ABD | POC | Nasal swab/nasopharyngeal swab | 80 | 29 | 24 | 7 |
| Zhang et al. (52) | China | CT | Unspecified | 60 | 60 | 48 | 8 |
| Shi et al. (53) | China | CT | Unspecified | 81 | 81 | 66 | 8 |
| Cheng et al. (54) | China | CT | Unspecified | 53 | 11 | 11 | 9 |
| Elslande et al. (55) | Belgium | ELISA | Blood | 103 | 94 | 86 | 11 |
| Huang et al. (56) | China | ELISA | Throat swab | 82 | 63 | 55 | 9 |
| Li et al. (57) | China | POC | Nasopharyngeal swab | 525 | 397 | 256 | 8 |
4.2. Characteristics of Studies
Table 3 shows the distribution of the performed tests by country. In the studies selected for the meta-analysis, no description was provided on the sample type in 2,442 cases. However, blood and throat swabs were the most commonly used sample types in the reviewed studies. Table 4 shows the sample distribution of the performed tests on 6,305 clinical cases. In the qualitative review, the studies were scored within the range of 4 - 12, and the mean qualitative score of the studies was estimated at 8.16 ± 1.84 (Table 2).
| Country | Test Methods | |||||
|---|---|---|---|---|---|---|
| POC | ELISA | CT | ||||
| No. of Studies | No. of Studies | No. of Studies | No. of Patients | No. of Studies | No. of Patients | |
| China | 2 | 102 | 9 | 1425 | 14 | 1671 |
| ABD | 3 | 159 | 1 | 40 | 1 | 141 |
| France | 0 | 0 | 0 | 0 | 1 | 572 |
| Italy | 2 | 111 | 1 | 64 | 1 | 462 |
| Jordan | 0 | 0 | 0 | 0 | 1 | 88 |
| Saudi Arabia | 0 | 0 | 0 | 0 | 1 | 150 |
| Brazil | 0 | 0 | 0 | 0 | 1 | 78 |
| Germany | 0 | 0 | 0 | 0 | 1 | 64 |
| Belgium | 0 | 0 | 1 | 94 | 0 | 0 |
| UAE | 1 | 499 | 0 | 0 | 0 | 0 |
| India | 1 | 72 | 0 | 0 | 1 | 147 |
| Spain | 1 | 55 | 0 | 0 | 0 | 0 |
| Japan | 1 | 112 | 0 | 0 | 1 | 112 |
| Canada | 1 | 42 | 0 | 0 | 0 | 0 |
| Pakistan | 1 | 45 | 0 | 0 | 0 | 0 |
| Total | 13 | 1197 | 12 | 1623 | 23 | 3485 |
Abbreviations: POC, point of care; ELISA, enzyme linked immunosorbent assay; CT, computer tomography.
| Sample Type | POC | ELISA | CT | Total | ||||
|---|---|---|---|---|---|---|---|---|
| NS | NP | NS | NP | NS | NP | NS | NP | |
| Unspecified | 0 | 0 | 0 | 0 | 17 | 2442 | 17 | 2442 |
| Blood | 4 | 278 | 8 | 1193 | 1 | 112 | 13 | 1583 |
| Throat swab | 0 | 0 | 1 | 63 | 2 | 652 | 3 | 715 |
| Nasal swab | 1 | 499 | 0 | 0 | 1 | 141 | 2 | 640 |
| Nasopharyngeal swab | 7 | 330 | 1 | 64 | 2 | 138 | 10 | 532 |
| Oral swab | 0 | 0 | 1 | 276 | 0 | 0 | 1 | 276 |
| Pharingeal swab | 1 | 90 | 0 | 0 | 0 | 0 | 1 | 90 |
| Saliva-oral swab | 0 | 0 | 1 | 27 | 0 | 0 | 1 | 27 |
Abbreviations: NS, number of studies; NP, number of patients; POC, point of care; ELISA, enzyme linked immunosorbent assay; CT, computer tomography.
4.3. Main Findings
The pooled sensitivity of POC, ELISA, and CT was estimated at 68.62%, 88.05%, and 75.43%, respectively. The mean true positive (TP) rate of POC, ELISA, and CT was reported to be 68.61%, 88.04%, and 79.25%, respectively. In addition, the mean false negative (FN) rate of POC, ELISA, and CT was calculated to be 31.38%, 11.95%, and 20%, respectively.
No significant difference was observed between the sample type and test sensitivity in one-way ANOVA (P > 0.05). On the other hand, a significant difference was observed between the applied method and test sensitivity (P < 0.05). A significant difference was also denoted between the country and test sensitivity (P < 0.05). According to Tukey’s post-hoc test, the sensitivity of ELISA and the tests performed was higher in China based on country (Table 5). However, no significant difference was observed between the sensitivity of TP in the paired-samples t-test (P > 0.05). A statistically significant difference was found between FN and test sensitivity. (P < 0.05) A statistically significant difference was found between FN (%) and test sensitivity. (P < 0.05) (Table 6)
Abbreviations: SD, standard deviation; TP, true positive; FN, false negative.
a P < 0.05
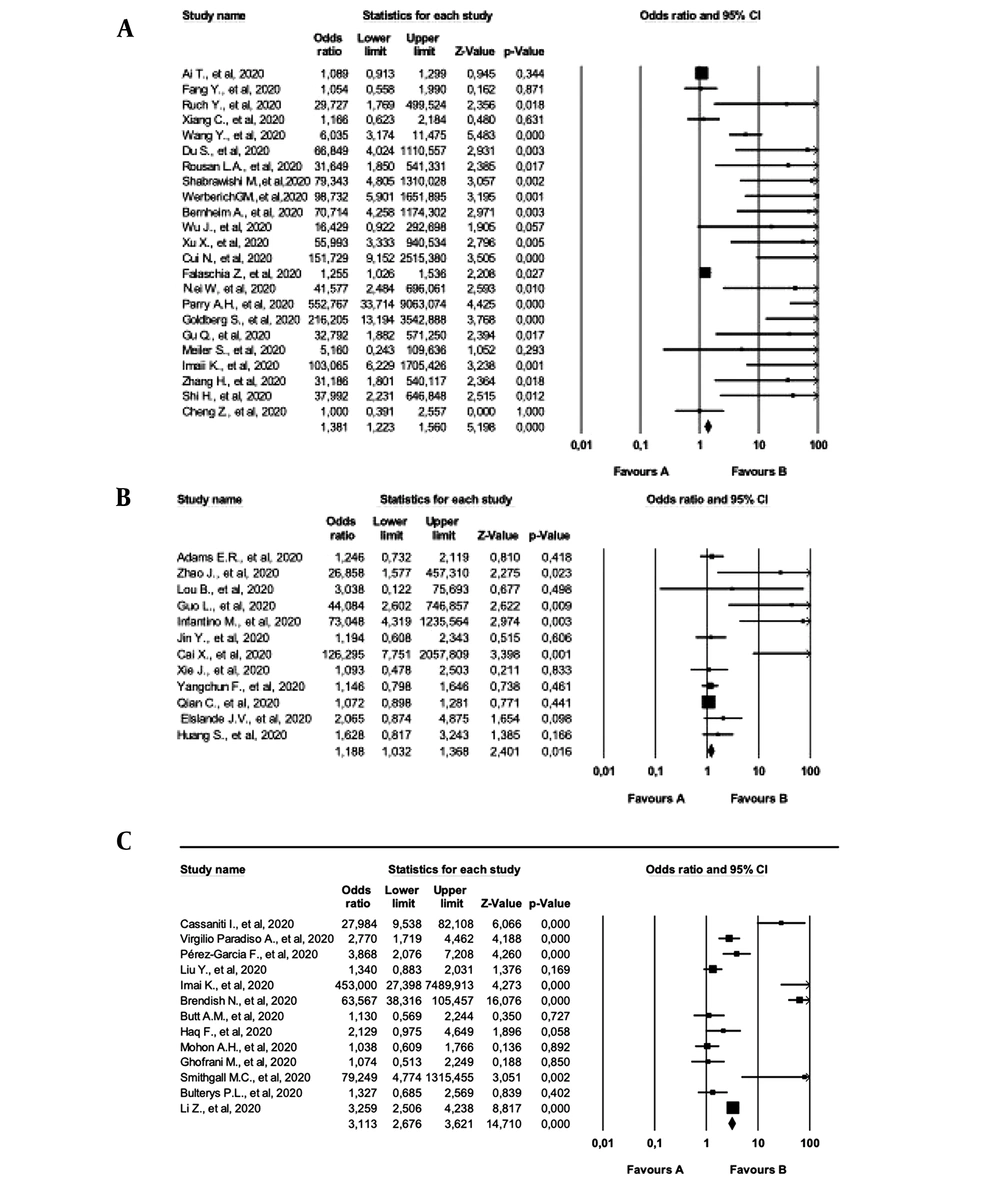
The random-effect (REX) and fixed-effect (FEX) models were used to calculate the effect size of the studies. According to the effect size analysis performed at 95% confidence interval (CI), ELISA and CT studies showed substantial heterogeneity (I2=67.5% and I2=85.4%, respectively). In addition, POC tests showed considerable heterogeneity (I2=94.8%), which was even higher than the I2 value of the effect size of all the tests (90.2%). Table 7 shows more statistical data obtained by the FEX and REX models.
| Model | No. | MES | Z | 95% CI | SD | Q | I2 | P-Value | |
|---|---|---|---|---|---|---|---|---|---|
| LL | UP | ||||||||
| POC | |||||||||
| FEX | 13 | 2.900 | 11.524 | 2.419 | 3.475 | NA | NA | NA | NA |
| REX | 13 | 4.074 | 3.261 | 1.751 | 9.478 | 12 | 232.050 | 94.829 | 0.000 |
| ELISA | |||||||||
| FEX | 12 | 1.188 | 2.401 | 1.032 | 1.368 | NA | NA | NA | NA |
| REX | 12 | 1.673 | 2.615 | 1.138 | 2.461 | 11 | 33.897 | 67.549 | 0.000 |
| CT | |||||||||
| FEX | 23 | 1.381 | 5.198 | 1.223 | 1.560 | NA | NA | NA | NA |
| REX | 23 | 7.963 | 7.423 | 4.604 | 13.772 | 22 | 150.752 | 85.407 | 0.000 |
| All studies | |||||||||
| FEX | 47 | 1.528 | 10.129 | 1.408 | 1.659 | NA | NA | NA | NA |
| REX | 47 | 4.391 | 8.447 | 3.115 | 6.190 | 47 | 479.700 | 90.202 | 0.000 |
Abbreviations: MES, mean effect size; REX, random effect; FEX, fixed effect; LL, lover limit; UL, upper limit; NA, non-Applicable.
In the present study, the REX model was used to calculate the effect size of the selected studies, and a higher effect size was observed in the CT group with the REX model. In the REX model, the effect size was calculated to be 4.391 at 95% CI. According to Cohen's classification, the reviewed studies had high effect sizes. (Table 7). Figure 2 depicts each of the reviewed studies in the forest plot chart.
5. Discussion
According to the results of the current meta-analysis, the existing evidence regarding the diagnostic accuracy of POC, ELISA, and CT for COVID-19 is characterized by limitations such as high bias risks and heterogeneity. Furthermore, our findings indicated that the sensitivity of CT and POC was consistently lower compared to ELISA. The sensitivity of POC has been estimated at 68.62%, indicating a significant weakness in this test compared to marketed bedside tests for COVID-19 diagnosis.
In the present study, a significant difference was observed between the sensitivity of the tests performed in China and the sensitivity of the tests conducted in Italy. Although no significant difference was denoted between the tests in China and other countries, the sensitivity of the tests performed in China was higher compared to other countries.
In a study aiming to evaluate the ELISA kits that are commercially produced for SARS-CoV2 without completed validation or those about to be validated, the sensitivity and differences of IgA and IgG antibody titers in patient sera were determined. According to the findings, this ELISA kit has been designed to bind to the S1 protein, and the results of IgA in the course of the disease were significantly correlated with the kit (8). Furthermore, the antibody in the kit is 90% similar to the other coronavirus N protein, indicating sufficient results and confirming the results of the serum test performed within five days from the onset of the infection by RT-PCR (58). Notably, evaluation of different recommended parameters for the course of the disease are essential to determining the specificity of SARS-CoV2 serological tests in the infection follow-up (59).
The benefits of POC tests are cost-efficiency, rapid implementation, and accurate results. However, the low sensitivity of this method is alarming. According to the studies in this meta-analysis, the sensitivity of the POC tests performed with serum/plasma samples is lower than the POC tests performed with nasopharyngeal samples; notably, the sample type should be considered in this test. Few patients are tested for suspected SARS-CoV-2 infection, and the overall results of our study may not be generalizable to all patients with suspected COVID-19.
Chest CT played a key role in the diagnosis of COVID-19 at the beginning of the outbreak and during the peak periods of the pandemic. Although RT-PCR is considered the ‘gold standard’ in this regard, the frequency of FN results after the first test and the absence of a laboratory kit in the early stages of the pandemic limit the early diagnosis of COVID-19 by this method. According to our findings, the tests used for the early diagnosis of COVID-19 (especially CT) have higher sensitivity in the regions that were first and heavily affected by the pandemic, such as China. Therefore, diagnostic criteria based on typical CT imaging features are only temporarily included in the current diagnostic-treatment guidelines in Hubei Province of China (60). These guidelines enable the early clinical diagnosis of COVID-19 due to the insufficiency of high-sensitivity tests such as ELISA and CT, as well as the RT-PCR diagnostic kits that contribute to the effective control of the current pandemic. The difference in this regard has several reasons, such as differences in CT sensitivity, heterogeneity in radiologists’ gaze, disease severity, and symptom onset.
Although RT-PCR is accepted as the ‘gold standard’, FN results are also encountered. According to Xie et al., more than 5% of COVID-19 patients initially had FN RT-PCR results, which became positive after multiple tests (61). However, 86% of these patients had positive chest CT images before their first negative RT-PCR results. Several cases of this issue are reported increasingly (60). Given the concerns about FN results, the limitations of RT-PCR, and the continued increase in the global cases of COVID-19, the British Thoracic Imaging Association highlights the importance of radiographic evaluation, especially in case of diagnostic uncertainty (62).
According to the CMA analysis in the current research, the I2 value was 90.20%, which indicated the high heterogeneity of the selected studies for the meta-analysis (P < 0.001). The main reasons of the heterogeneity between the studies are the diverse number of the evaluated patients, different demographic and clinical characteristics of the patients, differences in the applied methods, mistakes of the health personnel who evaluated the diagnosis tests, differences in the tested variables in ELISA (IgG, IgM, and IgG + IgM), differences in the commercial brand of POC and ELISA kits, differences in the virus structure in immunoserological tests (surface protein, nucleocapsid protein, surface/nucleocapsid proteins), and the interval between the onset of symptoms and performing CT, ELISA, and POC.
5.1. Limitations of the Study
The main limitations of our meta-analysis were the small sample size and exclusion of the studies evaluating fewer than 50 patients. In addition, meta-regression analysis was not performed since many studies would have to be excluded due to not reporting common variables. Another limitation was that we did not search databases such as OVID, Medline, and Embase, and some published studies might have been overlooked.
5.2. Conclusions
This is the first meta-analysis to simultaneously evaluate POC, ELISA, and CT sensitivity in COVID-19 diagnosis. According to the results, these tests had different sensitivity and specificity. ELISA was considered to be a more accurate diagnostic test for COVID-19 compared to POC and CT owing to its high sensitivity and true positivity rate, low FN rate, short processing time, and simple study procedure. Although helpful in diagnosis, confirmation of ELISA results by PCR remains the ‘gold standard’. Further investigations are urgently required regarding the diagnostic performance of the test methods used worldwide for COVID-19 on larger sample sizes and in greater detail.