1. Background
Polymyxins are antimicrobials used to treat infections caused by problematic microorganisms such as Pseudomonas aeruginosa, Acinetobacter baumannii, and carbapenemase-producing enteric bacteria that develop multi-drug resistance (1). Polymyxins were first produced in 1947 and used between the 1950s and 1980s. Later, their nephrotoxic side effects were noticed, and they were not preferred for many years, except for the treatment of cystic fibrosis. However, infections caused by resistant bacteria have been increasing in recent years. The failure of treatment with current antibiotics has led to the re-thinkable and preference of polymyxins in the treatment options (2, 3).
Polymyxins are synthesized out of ribosomes by Paenibacillus polymyxa, which chemically consists of five different compounds (polymyxin A - E). Only polymyxin B and polymyxin E (colistin) are used in the clinic (4, 5), and colistin targets the bacterial cell membrane. Colistin, a cationic peptide, binds to the anionic lipopolysaccharides found in the outer membrane of gram-negative bacteria. Displacing divalent cations (Ca2+, Mg2+) that keep lipopolysaccharide molecules together causes the death of bacteria because of the deterioration of the outer membrane and increased permeability (5-7). Besides its antibacterial properties, colistin inhibits endotoxin production by binding to the lipid A portion of lipopolysaccharides. The sensitivity of bacteria to colistin is related to the amount of phospholipid contained in the cell membrane and the level of divalent cations in the medium (5). Colistin is an antibiotic used as a last option in treating gram-negative bacteria with multi-drug and carbapenem resistance in recent years (8). Increased colistin resistance has been observed due to excessive and inappropriate use. The polymyxin resistance mechanism reported until the end of 2015 is chromosomally derived and generally causes mutations in genes encoding specific two-component regulatory systems (9).
However, in 2015, plasmid-mediated colistin resistance and the mobile colistin resistance (mcr) gene mcr-1 were reported for the first time, which caused colistin resistance by adding phosphoethanolamine to the lipid A region of the membrane (10). In July 2016, the plasmid-mediated colistin resistance gene mcr-2 was detected in Belgium’s Escherichia coli isolates, mainly isolated from pigs (11). The mcr-2 is a phosphoethanolamine transferase that probably modifies the limb of lipopolysaccharide. The amino acid identity of mcr-2 is 80.6%, like that of mcr-1 (12). Following the initial findings, many recent studies have identified different variants of mcr-1 and eight new mcr genes (from mcr-3 to mcr-10) (13-23). Detection of plasmid-mediated colistin resistance genes is essential for understanding colistin resistance mechanisms.
2. Objectives
This study aimed to evaluate the presence of plasmid-mediated colistin-resistance genes mcr-1 and mcr-2 in colistin-resistant enteric gram-negative bacilli obtained from various patient samples.
3. Methods
3.1. Bacterial Isolates, Identification, and Susceptibility Testing
Enterobacterales isolated from various samples sent to Ondokuz Mayıs University Faculty of Medicine Microbiology Laboratory in 2016 were included. Vitek MS (bioMeriux, France) was used for bacterial identification, and Vitek 2 Compact (bioMeriux, France) automated system was used to determine colistin resistance. Colistin resistance was confirmed by the broth microdilution method. The isolates were stored at -20°C until molecular analysis.
3.2. Molecular Analysis
DNA of colistin-resistant isolates was extracted by boiling method. The 309 bp fragment of the mcr-1 gene and 567 bp fragment of the mcr-2 gene was amplified with specific primers in all colistin-resistant Enterobacterales isolates to detect the plasmid-mediated colistin resistance genes (Table 1). The used primers were determined after a literature search (14, 15). Multiplex polymerase chain reaction (PCR) was performed under the following conditions: Heat denaturation for 15 minutes at 94°C, 25 cycles of the 30s at 94°C, 90s at 59°C, and 1 min at 72°C, and a final extension step of 10 min at 72°C. The PCR products were applied to a 2% agarose gel. Subsequently, the DNA bands of the isolates were compared with 100 bp DNA markers and examined on the imaging instrument.
| Genes | Primers Sequences (5’-3’) | Amplicon Size (bp) |
|---|---|---|
| mcr-1 | 309 | |
| CLR5-F | CGGTCAGTCCGTTTGTTC | |
| CLR5-R | CTTGGTCGGTCTGTAGGG | |
| mcr-2 | 567 | |
| mcr-2-F | TGTTGCTTGTGCCGATTGGA | |
| mcr-2-R | AGATGGTATTGTTGGTTGCTG |
4. Results
The most frequently isolated bacteria were Klebsiella pneumoniae (37.6%) and Serratia marcescens (31.7%) in colistin-resistant 170 Enterobacterales isolates obtained from clinical samples. Table 2 shows the distribution of isolated bacteria by species. The isolates were most frequently isolated from the samples sent from internal medicine (44.7%) and neurology (13.5%) clinics. Table 3 presents the distribution of samples based on clinics, the most frequent of which are blood samples (30%). Table 4 indicates the distribution of the models.
| Bacteria Species | No. (%) |
|---|---|
| Klebsiella pneumoniae | 64 (37.6) |
| Serratia marcescens | 54 (31.7) |
| Proteus mirabilis | 23 (13.5) |
| Morganella morganii | 13 (7.6) |
| Providencia rettgeri | 10 (6) |
| Proteus vulgaris | 2 (1.2) |
| Escherichia coli | 2 (1.2) |
| Providencia stuartii | 1 (0.6) |
| Serratia liquefaciens | 1 (0.6) |
| Clinics | No. (%) |
|---|---|
| Internal medicine | 76 (44.7) |
| Neurology | 23 (13.5) |
| Surgical services | 15 (8.9) |
| Pediatrics | 12 (7) |
| Infection | 12 (7) |
| Chest diseases | 9 (5.3) |
| Emergency and first aid | 6 (3.5) |
| Orthopedics and traumatology | 6 (3.5) |
| Urology | 4 (2.4) |
| Dermatology | 4 (2.4) |
| Otolaryngology | 1 (0.6) |
| Gynecology and obstetrics | 1 (0.6) |
| Cardiology | 1 (0.6) |
| Samples | No. (%) |
|---|---|
| Blood | 51 (30) |
| Exuda | 43 (25.3) |
| Tracheal aspirate | 33 (19.4) |
| Sterile body fluid | 16 (9.4) |
| Urine | 14 (8.2) |
| Sputum | 11 (6.5) |
| Cerebrospinal fluid | 2 (1.2) |
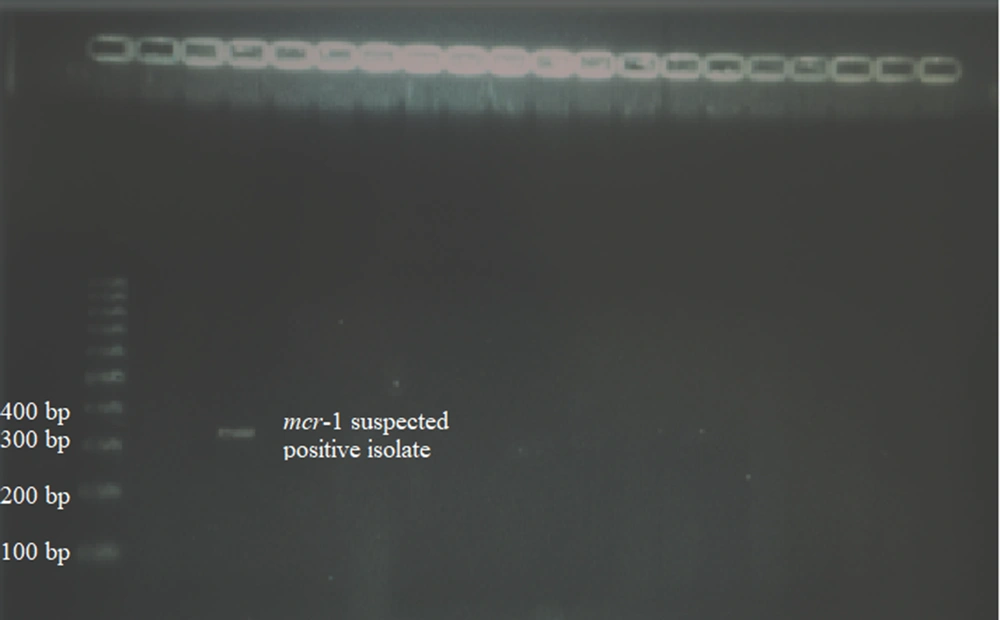
As a result of the molecular process, an isolate forming a band in the 309 bp region was determined to be mcr-1 suspected positive. The PCR gel image of this isolate is given in Figure 1. No band-forming isolate was found in the 567 bp region for the mcr-2 gene. Sequence analysis was applied to the PCR products of the mcr-1 suspected positive strain, and the suspected isolate was determined to be mcr-1 negative at the end of the sequence analysis.
5. Discussion
Colistin is a polymyxin group antibiotic used as a last resort in treating gram-negative bacteria, especially carbapenem-resistant Enterobacterales (5, 24). Colistin has been reported by the World Health Organization (WHO) as one of the ‘highest priority critically important antimicrobials’ (25). The increasing prevalence of CRE worldwide has led to the widespread use of colistin and increased colistin resistance, especially in countries where CRE is endemic, such as Italy and Greece (26-30).
Colistin resistance mechanisms are mostly chromosomal and usually cause mutations in genes encoding specific bicomponent regulatory systems. The mcr-1 gene, which caused plasmid-mediated colistin resistance, was first identified in 2015 in E. coli isolates, and the identification of the mcr-2 gene followed it in 2016 (10, 11). The rapid spread of plasmid-mediated resistance mechanisms between species by horizontal gene transfer makes this resistance feature significant clinically and epidemiologically due to the rapid increase in the frequency of colistin-resistant isolates and the potential of these isolates to spread and cause epidemics (8, 31).
In studies performed with colistin-resistant isolates, most of the isolates were K. pneumonia (8, 32). Similarly, the most frequently isolated species in the present research was Klebsiella spp. (37.6 %) among colistin-resistant Enterobacterales isolates. After the first report for mcr-1, microbiologists have reported the presence of mcr-1 in Enterobacterales isolates from chicken meat, E. coli isolates from clinical samples and animals, and ESBL-producing K. pneumoniae isolated from clinical samples in different countries (33-35).
mcr-1 was first reported by Kurekci et al. in chicken meat in Turkey (36). Subsequently, studies in which mcr-1 was detected in clinical isolates have been reported (37-39).
The number of studies investigating the mcr-2 gene is less than the mcr-1 gene. Xavier et al. reported that they first detected the mcr-2 gene in colistin-resistant E. coli isolates isolated from pigs and cattle in Belgium (11). Liassine et al. detected the mcr-2 gene for the first time in colistin-resistant Enterobacterales isolated from a clinical sample (12). As far as we know, mcr-2 has not yet been detected in Turkey.
In this study, mcr-1 and mcr-2 genes were not detected. Similar to the present research, Sari et al. and Hosbul et al. could not detect mcr-1 and mcr-2 genes in clinical isolates (8, 40).
5.1. Conclusions
Since 2014, the colistin resistance level has increased, and the worrying discovery of mcr genes requires urgent precautions, even though this study did not detect mcr-1 and mcr-2. Our isolates date back to 2016, which is one of the study’s limitations. New studies investigating mcr genes in new isolates are needed to understand the mechanism of resistance and identify resistant isolates.