1. Background
The esophagus is the first food passage to the body after the mouth, which can be affected by various disorders, such as cancer in the long term (1). It is seen in two forms of esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC), depending on the involved cells, with the rate of higher mortality in worldwide (2, 3). The environmental factors including smoking, alcohol drinking, hot liquid drinking, meat, obesity, and gastroesophageal reflux are considered as the higher risk factors. In addition, studies have indicated that the genetic factors are also involved in the development and progression of the ESCC (4, 5).
Identification of prognostic and diagnostic biomarkers is one of the effective and crucial requirements for clinical evaluation and treatment management (6). To date, it has been indicated that nucleic acids in the plasma, serum, and tissue cancer patients, which can be used as a diagnostic tool. Accordingly, previous studies have conducted on noncoding RNA as potential tumor biomarkers for prognosis and diagnosis of cancer.
However, in recent years, diagnostic tools of plasma long non-coding RNAs (lncRNAs) have been investigated in a variety of cancers, especially in ESCC (7).
Several investigations on lncRNA have shown that these molecules play a crucial role in regulating tumor suppressor genes. They are also involved in the progression and development of various cancers (8).
Various lncRNAs have been investigated the potential roles in the pathogenesis of ESCC including lncRNA-POU3F3(7), lncRNA-BANCER (9), lncRNA-CCAT-1 (10), and lncRNA-H19 (11). Recently, the findings revealed that the lncRNA-RP11_766N7.4 gene was up-regulated in the ESCC tissues (12). An investigation has shown that the over-expression of lncRNA-RP11_766N7.4 gene promotes the inhibition of cellular metastasis and invasion of ESCC cells by inhibiting the epithelial-mesenchymal transition (EMT) process (12). LncRNA-LINC02389 gene was expressed in various tissues of the body, including the lung, stomach, kidney, and heart (13). This gene is located on chromosome 12q14.3 containing 3 exons. The gene expression change of this lncRNA has not been evaluated in ESCC samples.
2. Objectives
The aim of this investigation was to evaluate the lncRNA-LINC02389 gene expression variation and clinical significance in the progression of ESCC.
3. Methods
3.1. Sampling the Patients
In our preliminary study was performed on seventy-five paired samples of paraffin block containing 75 cancerous and 75 marginal non-cancerous tissues. Samples were taken from patients with ESCC by endoscopy, referred to the International Hospital of the Tabriz University of Medical Sciences, whose malignancy was diagnosed and confirmed by pathologists. In this study, the subjects were selected among those who met the definite malignancy diagnosis by histopathologists and determined stage type (I-II-III-IV) of the ESCC. The exclusion criteria involved those who had undergone chemotherapy or radiotherapy. The taken samples were transferred to the laboratory as paraffin and kept at -70°C until the RNA extraction. In order to extract total RNA, four slices of 10µm paraffin block samples were cut through microtome for deparaffinization.
The protocols confirmed by the Ahar Branch Islamic Azad University and signed informed consent and questionnaires were received from each case.
All RNA extraction procedures were performed according to the miRcuRY ™ RNA Isolation kit of FFPE kit (Exiqon’s- Denmark). To determined qualitative and quantitative of RNA extracted, we have used agarose gel electrophoresis and UV spectrophotometer at 260/280 nm (using the NanoDropTM ND-1,000, NanoDrop Technology, Wilmington, DE, USA), respectively. Extracted RNAs were stored at -80°C until cDNA was synthesized. In order to synthesize cDNA from extracted RNAs, the Revert AID TM Firs Standard cDNA Synthesis kit (Fermentas, Germany) was used.
3.2. Real Time PCR Method
In order to design the required sequences primer, Gene runner software was used and its specificity was examined through BLAST software. To amplification of the lncRNA-LINC02389 gene used the following sequence: forward, 5´-CAGAACGCAATGGAAACAGA-3´and reverse, 5´-GATGATGCCCAGAGGAAGAG-3´. The real-time PCR reaction was performed with Rotter Gene Q device (Qiagen-USA) in duplication to ensure the accuracy of amplification. In this study, the miR-U6 was used as a housekeeping gene to correct the errors caused by sampling and normalization of the reaction. Real-time PCR reaction was performed for diluted cDNAs with a ratio of 1 to 50 and at a final volume of 15 µL. The components of reaction mixture included 9 µL Master Mix cyber green, 1 µL cDNA, 1 µL of each of the forward and reverse primers of lncRNA-LINC02389 and miR-U6 genes and 1 µL of deionized distilled water. Samples were prepared in 40 cycles of three-stages, including initial denaturation (at 95°C for 40 seconds), annealing (at 60°C for 350 seconds) and extension (at 72°C for 60 seconds). To measure the relative gene expression change, we were applied to the 2-ΔΔct method. PCR reaction was carried out with housekeeping gene primers to validate the cDNA synthesized.
3.3. Statistical Analysis
The Paired-Samples t-test was applied to examine the significant difference of gene expression variations in cancer samples in comparison with the marginal samples. Moreover, the normal distribution of gene expression in the samples was evaluated using the Kolmogorov-Smirnov test (K.S). Pearson correlation test was used to examine the correlation between gene expression changes and clinical features. In this study, a significant statistical difference of P < 0.05 was considered for all cases.
4. Results
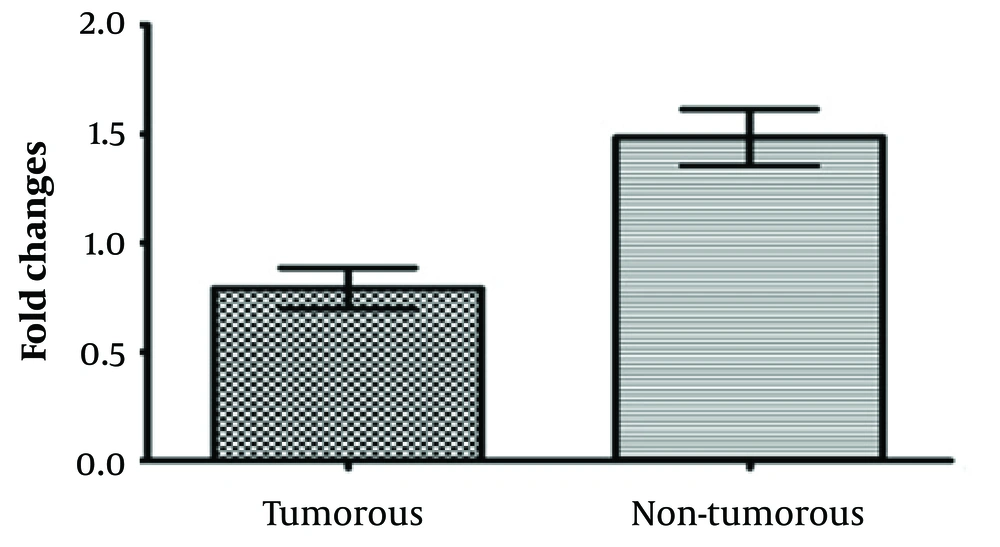
A total of 150 paraffin samples (75 esophagus cancer samples and 75 marginal tissue samples) were evaluated to determine the lncRNA-LINC02389 gene expression variations. The rate of changes in the expression of lncRNA-LINC02389 gene was measured using the real-time PCR method. The single peak of the lncRNA-LINC02389 gene melting curve indicated the absence of a non-specific product or a dimer primer.
Our findings revealed that the lncRNA-LINC02389 gene was significantly decreased in the tumorous tissue compared to the marginal non-tumorous tissues (P < 0.05) (Figure 1).
In addition, the results showed a significant relationship between the rate of lncRNA-LINC02389 gene expression variations and tumor differentiation (P = 0.003), but there was no relationship between lncRNA-LINC02389 gene expression and other tumor characteristics (Table 1).
| Parameter | No. | Mean ± SD | P-Value |
|---|---|---|---|
| Tumor differentiation | |||
| Well and moderate | 55 | 0.860 ± 0.169 | 0.003 |
| Poor | 20 | 1.7480 ± 0.217 | |
| Tumor stage | |||
| I+II | 37 | 1.010 ± 0.315 | 0.151 |
| III+IV | 38 | 1.961 ± 0.219 |
There was a statistically significant relationship between the lncRNA-LINC02389 gene expression changes and demographic parameters such as alcohol drinking (P = 0.010) and drinking hot liquids (P = 0.002) (Table 2).
| Parameters | No. (n = 75) | lncRNA-LINC02389 Gene Expression | P-Value | |
|---|---|---|---|---|
| Low | High | |||
| Drinking hot liquids | ||||
| Yes | 46 | 11.0 | 35.0 | 0.002 |
| No | 29 | 17.0 | 12.0 | |
| Smoking | ||||
| Heavy | 40 | 18.0 | 22.0 | 0.345 |
| Light | 35 | 12.0 | 23.0 | |
| Alcohol drinking | ||||
| Yes | 41 | 7.0 | 34.0 | 0.010 |
| No | 34 | 15.0 | 19.0 | |
| Socioeconomic status | ||||
| Good + Borderline | 57 | 31.0 | 26.0 | 0.252 |
| Poor | 18 | 7.0 | 11.0 | |
| Stage | ||||
| I and II | 37 | 13.0 | 23.0 | 0.151 |
| III and IV | 38 | 8.0 | 30.0 | |
| Overall five-year survival rate | ||||
| Positive | 34 | 12.0 | 22.0 | 0.153 |
| Negative | 41 | 9.0 | 32.0 | |
5. Discussion
Esophageal carcinoma is one of the malignant gastrointestinal cancer types, which were highly released and metastasis in the adjacent lymphoid tissues and glands (6). Despite the developments and progress in screening, diagnosing and cancer treating, the prognosis of esophageal cancer is still poor and its 5-year survival rate is very low (12). One of the main goals of the researchers was to find specialized diagnostic biomarkers to cancer diagnosed in the early stages. In recent years, the role of lncRNAs in the early diagnosis of various cancers has drawn the attention of many researchers. Based on the previous investigation conducted on lncRNAs, the role of these molecules in the cell proliferation, differentiation, and metastasis of ESCC has been recognized (9). The gene expression change of lncRNA-LINC02389 has not been evaluated in ESCC samples. The aim of this investigation was to evaluate the lncRNA-LINC02389 gene expression variation and clinical significance in the progression of ESCC.
This investigation is a preliminary study regarding the potential roles of lncRNA-LINC02389 gene expression variations in the pathogenesis of ESCC. Our data disclosed that the lncRNA-LINC02389 gene was significantly down-regulated in the tumorous tissue compared to the marginal non-tumorous ESCC samples.
The findings revealed that a significant relationship between lncRNA-LINC02389 gene expression variations and tumor differentiation. In addition, results showed a significant relationship between the lncRNA-LINC02389 gene expression changes and life style.
To date, several surveys assessed the possible roles of lncRNAs gene expression variations in ESCC progression, which were showed contradictory results.
Tong et al. have been examined several lncRNAs gene expression changes in cancerous tissues and circulating in the plasma of patients with esophagus cancer (7). The results showed that the plasma levels of lncRNA-POU3F3, lncRNA-HNF1A-AS1, and lncRNA-SPRY4-IT1 in ESCC patients were higher than those in the control group. Among the three cases of lncRNA investigated, the plasma level of lncRNA-POU3F3 showed the highest diagnosis rate for ESCC. In addition, the combined use of the lncRNA-POU3F3 and SCCA could provide more effective diagnostic performance for esophagus cancer. Most importantly, this combination was effective in the diagnosis of ESCC in the early stages (7).
An investigation by Chen et al. in the lncRNA-HOTAIR gene expression variation showed that the expression level of lncRNA in esophagus cancerous tissues was significantly higher than that of the adjacent tissues and had a positive correlation with tumor node metastasis staging (TNM). They also disclosed that the lncRNA-HOTAIR gene silencing in an esophageal squamous cancer cell line (KYSE30) inhibited the invasion of cancer cells, while it increased the response of the cells to apoptosis (14). Accordingly, the results of the present study showed a significant relationship between the lncRNA-LINC02389 gene expression with the stage.
The results study conducted by Huang et al. on the relationship between the lncRNA-HOTAIR gene expression and possible roles of pathogenesis in esophagus cancer, showed that the lncRNA-HOTAIR gene expression was significantly increased in cancerous comparison with normal cells. Moreover, they were revealed that a significant relationship with tumor size and metastasis. In contrast, down-regulation of the lncRNA-HOTAIR inhibited the cell proliferation and invasion of esophagus cancer cells. As a result, lncRNA-HOTAIR played a major role in the development of cancer, which could be used as a clinical prognostic marker for ESCC (11).
In addition, Yang et al. have assessed the lncRNA-HNF1A-AS1 gene expression changes in ESCC and the findings revealed that significantly higher expression levels in the early stages of esophagus cancerous cells compared with the normal tissue. They have also revealed that the lncRNA-HNF1A-AS1 gene silencing inhibited the cell proliferation and progression, suppressed the S-phase of the cell cycle, and inhibited the migration and cellular invasion of cancerous cells. It was also found that the lncRNA-HNF1A-AS1 gene silencing inhibited the activity of the lncRNA-H19 gene, showing a positive correlation between two types of lncRNA gene expression in the early stages of ESCC (15).
Gao et al. surveyed the role of lncRNA-H19 in regulating the H19 imprinting control region (ICR) and Insulin-like growth factor 2 (IGF-2) expressions and its association with ESCC progression. The results showed that the lncRNA-H19 was significantly down-regulated in ESCC with high invasion and larger tumor size, which leads to over-expression of IGF2 in tumor progression. In addition, they stated that reducing the lncRNA-H19 expression could potentially lead to the identification of people at the risk of ESCC development and progression (16).
Previously, the findings investigation was disclosed that the lncRNA-CASC9 was significantly over-expressed in esophagus tumorous tissues. The lncRNA-CASC9 gene silencing could be inhibited cells growth and proliferation and blocking the cell cycle in the G1/S phase. The reducing effect of lncRNA-CASC9 was due to the involvement in the binding of the EZH2 strengthening agent to the promoter of the EZH2 gene (negative regulation) and H3K27me3 in this region. The researchers stated that lncRNA-CASC9 could be used as a valuable marker for diagnosis and prognosis of ESCC (11).
Zhang et al. in their study showed that the expression of lncRNA-CCAT1 was significantly higher in the ESCC tissue. In addition, a significant relationship was also found between the level of lncRNA-CCAT1 and lymph node metastasis and the TNM stage (10) (Table 3).
| LncRNA | Size | Cytogenetic | Regulation | Biological Functions | References |
|---|---|---|---|---|---|
| LncRNA-H19 | 2.3 kb | 11p15.5 | Down-regulated | Loss of imprinting at the H19 locus resulted in high H19 expression in cancer of the esophagus | (11) |
| LncRNA-CCAT1 | 1.7 kb | 8q24.21 | Down-regulated | CCAT1 had a significant impact on ESCC cell proliferation | (10) |
| LncRNA-HOTAIR | 5.4 kb | 12q13.13 | Up-Regulation | Silences the tumor suppressor genes by interacting with EZH2 and enhancing H3K27me3 | (14) |
| LncRNA-POU3F3 | - | 2q12.1 | - | Promotes DNA methylation in esophageal squamous cell carcinoma cells | (7) |
| LncRNA-CASC9 | - | 8q21.13 | Up-regulation | Up-regulation of lncRNA CASC9 promotes esophageal squamous cell carcinoma growth by negatively regulating PDCD4 expression through EZH2 | (17) |
Another group of potential tumor biomarkers investigated in the ESCC including squamous cell carcinoma antigen (SCCA) (17), carcinoembryonic antigen (CEA) (18), cancer antigen 19-9 (CA19-9) (19), Matrix metalloproteinase (MMP-9) (20), interleukin-6 (IL-6) (21), CYFRA 21-1 (22), dickkopf WNT signaling pathway inhibitor 1 (DKK-1) (23), macrophage-colony-stimulating factor (M-CSF) (24), miR-18a (25), miR-1246 (26). As mentioned above, the study was designed as a preliminary investigation, which assessed the potential role of lncRNA-LINC02389 gene expression changes in pathogenesis of ESCC patients. There are several limitations in our study, which consisting of the small sample size of patients and used FFPE samples for gene expression analysis. This small number of patients may have led to the findings that lncRNA-LINC02389 expression level had no real impact on pathogenesis of ESCC. Also, the results showed that most of these molecules did not have adequate specificity and sensitivity to be used as biological markers. However, future studies are needed to clarify the clinical significance of this LncRNA expression level in other types of cancers. In addition, the molecular mechanism of the lncRNA-LINC02389 in tumor progression and development needs to be determined in the next study.
5.1. Conclusions
The results of the present study showed that lncRNA-LINC02389 may be acted as a tumor inhibitor in ESCC. These results provided a new insight to the development prognosis of ESCC. Moreover, the results indicated that lncRNA-LINC02389 could be a potential predictor marker and a therapeutic goal in ESCC. However, further investigations were required for clarity the potential roles of lncRNA-LINC02389 in cancerogenesis of ESCC.