1. Context
The increasing rate of nosocomial infections is one of the significant issues that educational medical centers and hospitals face (1). These infections are considered a severe risk factor threatening the health of almost all hospitalized patients (2). Appropriate use of disinfectants is an influential factor in preventing hospital infections (3). Disinfectants or antiseptic agents are a group of antimicrobial substances used on the surface of living tissues of the skin and body to destroy or prevent the growth of bacteria, viruses, fungi, and spores (4). Understanding the principles of sterilization and disinfection and applying the appropriate infection control measures can be very important in health issues (5). Nevertheless, the physical and chemical structure of these substances, their inappropriate use at nonstandard concentrations, and the poor structure of hospitals have contributed to the ineffectiveness of disinfectants against nosocomial pathogens like Escherichia coli, Pseudomonas aeruginosa, Klebsiella, Enterobacter, and Acinetobacter. In recent years, Staphylococcus aureus and Staphylococcus epidermidis have been prevalent (6). Consequently, nosocomial pathogens develop acquired resistance against common antibiotics and antiseptics used in hospitals, adding to these microorganisms' inherent resistance (7). Hence, resistance to antiseptics and disinfectants increasingly develops (8).
Extensive studies have been conducted in Iran to identify the prevalence of antibiotic-resistant genes in various bacteria. These studies have shown that the prevalence of nosocomial Infections in Iran is 0.6, and antibiotic resistance genes in bacteria are increasing (1). However, no comprehensive research provides all the results in one single study.
2. Objectives
This study aimed to comprehensively investigate the frequency of biocide resistance genes in bacterial isolates in Iran.
3. Methods
3.1. Search Strategy
The systematic review was performed at the infectious diseases department of Kermanshah University, Iran and Iranian and English databases including Danesh Gostar Barkat, Scientific Information Database Index Medicus, Irandoc, Magiran, Google Scholar, PubMed, MEDLINE, EMBASE Cochrane, CINAHL databases, Ovid, Scopus, ISI Web of Science, Cochrane Library MD Consult, and Science Direct were searched for relevant mesh terms related to the topic of the present study. The mesh keywords included Resistance genes, qacEΔ1, qacE, Antiseptic resistance genes, Frequency, gram-positive bacteria, gram-negative bacteria, and Iran.
3.2. Inclusion and Exclusion Criteria
Studies that investigated the frequency of resistance genes to antiseptics in bacterial isolates, whose full texts were available, were included in the study. However, the reports investigating the frequency of resistance genes to antiseptics in nonbacterial isolates were excluded from the study.
3.3. Extracting the Data
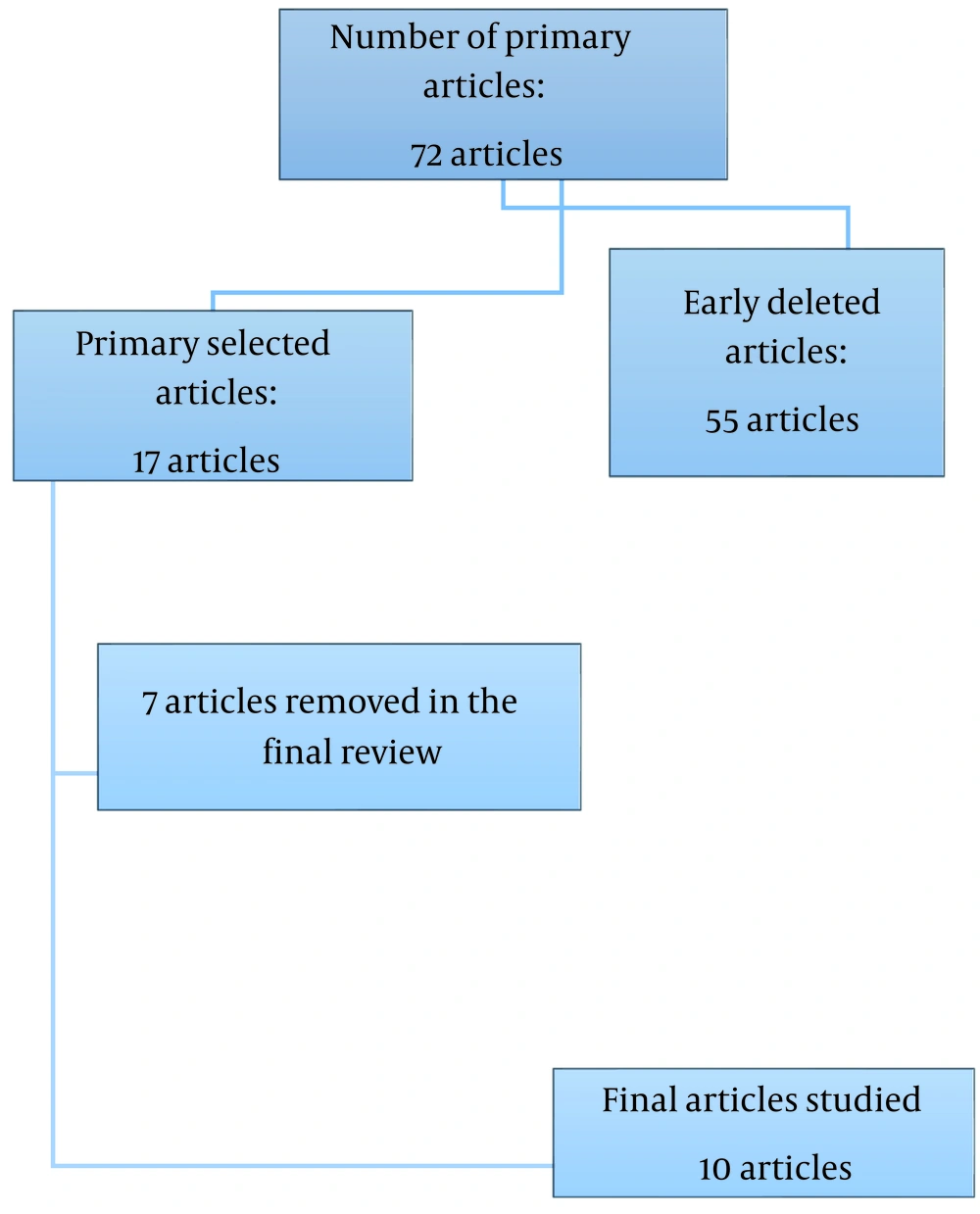
After searching the database, 72 studies were found, 55 of which did not have available full texts. Out of the remaining 17 studies, seven investigated the frequency of resistance genes to antiseptics in nonbacterial isolates. Therefore, data extraction and interpretation were done on ten studies, as shown in Figure 1.
The required information, such as the name of the first author, the year of the study, the location, the type of study, the age and gender of the patients, the type of samples examined, the species and genera of bacteria examined, and the frequency of antiseptic resistance genes, was extracted after collecting related articles. Then, the studies were categorized by the authors. In the end, the results were interpreted, and the final result was reported.
4. Results
After collecting the data, the results were briefly reported in Table 1. The findings showed that three were conducted in Tehran, two in Zahedan and Arak, and one in Shahrekord, Lorestan, and Qazvin, among the ten remnants included in the studies. Six studies investigated biocide resistance genes in gram-positive bacteria, and four investigated gram-negative bacteria. All the samples of these studies were collected from clinical samples. There were 975 bacterial isolates, including 516 gram-positive and 459 gram-negative bacteria. PCR method was used in all studies to identify the genes investigated. Among 516 Staphylococcus isolates, the highest frequency was related to the QacAB gene, with an average of 28.48%. The frequency of the SMR gene was reported as 19.8% on average. Among the gram-negative bacteria, the qacEΔ1 gene had the highest frequency, averaging 45% (Table 1).
| First Author | Year | Place | Sample | Studies Type | Method of Detection | Bacteria-Name | Gene-Name | Frequency | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Hadadi | 2019 | Arak | 150 | Clinical | PCR | Escherichia coli | QacE, QacF, QacG, qacEΔ1 | 0.0, 0.0, 0.0, 100 | (9) |
| Damavandi | 2017 | Shahr-e Kord | 120 | Clinical | PCR | Staphylococcus aureus | QacAB, QacC, SMR | 12.5, 21.7, 31.7 | (10) |
| Keshavarz-Hedayati | 2019 | Qazvin | 141 | Clinical | PCR | Acinetobacter baumannii | QacE, QacEΔ1 | 17.0, 59.0 | (11) |
| Saadatian | 2016 | Tehran | 83 | Clinical | PCR | Klebsiella | OqxAB, AcrAB | 96.0, 97.0 | (12) |
| Tahmasebi | 2017 | Zahedan | 89 | Clinical | PCR | Staphylococcus aureus | SMR, QacAB | 23.7, 49.5 | (13) |
| Azadpour | 2015 | Lorestan | 85 | Clinical | PCR | Klebsiella | QacEΔ1 | 30.6 | (14) |
| Taheri | 2016 | Arak | 165 | Clinical | PCR | Staphylococcus aureus | QacAB, SMR | 16.7, 6.7 | (15) |
| Dadook | 2014 | Tehran | 40 | Clinical | PCR | Staphylococcus aureus | SMR | 25.0 | (16) |
| Bokaeian | 2016 | Zahedan | 60 | Clinical | PCR | Staph coagulase | QacA, SMR | 52.7, 58.3 | (17) |
| Nowroozi | 2011 | Tehran | 42 | Clinical | PCR | Staphylococcus aureus | QacAB, SMR | 19.4, 45.2 | (18) |
The Main Characteristics Studies Included
5. Discussion
Disinfectants or antiseptics are regularly used for sterilizing or disinfecting medical equipment such as endoscopic devices, surgical equipment and dressings, operation, and delivery rooms, burn departments, syringes and serum sets, and corridor floors of hospitals. However, many of these materials lose their effectiveness due to their physical and chemical structure, inappropriate use, the lack of standardized effective concentrations, and the improper structure of hospitals (19). Nosocomial infections have caused significant problems in medical education centers. On the other hand, surgical wound infections account for 25% of all nosocomial infections and significantly contribute to improving the economic burden caused by treatment expenses of surgical complications, which are the leading causes of death in hospitalized patients (20). One of the essential factors in the spread of hospital infections is the improper use of disinfectants. Contamination of medical devices and equipment and the consequent development of infections like hospital-acquired pneumonia, urinary tract infection, and surgical wound infections can increase the length of hospitalization and mortality (21).
Nosocomial infections are a growing risk factor for bacterial resistance to antibiotics and antiseptics. Bacteria are the main pathogenic agents involved in nosocomial infections (22). In addition, they are the first agents to develop resistance to commonly used antibiotics and antiseptics (17). In general, gram-negative bacteria are less sensitive to biocides than gram-positive bacteria, and the reason for this is the outer membrane of these bacteria, which serves as a barrier to penetration of the disinfectant (23). The simultaneous emergence of resistance to biocides and antibiotics is one of the most concerning phenomena. One of the important characteristics of the responsible bacteria in nosocomial infections is the presence of a system that excretes biocidal compounds from inside to outside of the cell, which is one of the mechanisms of resistance of these bacteria to these compounds (24). Some bacteria may carry multidrug delivery systems such as qacEΔ1 and qacE proteins. The qacEΔ1 gene is a part of the conserved end of class 1 integrons (25). These integrons are usually isolated from antibiotic-resistant clinical strains of Enterobacteriaceae and Pseudomonas species. The qacEΔ1 gene is a more effective and active form of the qacE gene. This gene encodes resistance to biocides such as quaternary ammonium compounds (QACs) and dyes like ethidium bromide. The face gene is a part of the end of these genetic fragments in some bacteria (26, 27). Extensive studies have been conducted in Iran to identify the prevalence of genes resistant to antiseptics in different bacteria. Hence, the purpose of this research was to systematically review the frequency of genes resistant to biocides in bacterial isolates in the country. Of the ten studies selected in this research from different parts of Iran, six investigated biocide resistance genes in gram-positive bacteria, and four investigated the same gram-negative bacteria. There were 975 bacterial isolates, including 516 gram-positive and 459 gram-negative bacteria. The PCR method was used in all studies to identify the so-called genes. Among the 516 gram-positive isolates, the highest frequency was related to the QacAB gene, with an average of 28.48%, and the frequency of the SMR gene was reported as 19.8% on average. Among the gram-negative bacteria, the qacEΔ1 gene had the highest frequency, averaging 45%. In Helal and Khan on the investigation of the frequency of qacΔE1 and qacE genes and their relationship with resistance to antibiotics and antiseptics in Pseudomonas aeruginosa isolates, 34.4% of 136 isolates of Pseudomonas aeruginosa were multidrug resistant (MDR). qacΔE1 gene was found in 57.8% of MDR Pseudomonas strains and 21.4% of antibiotic-sensitive ones. Furthermore, qacE gene was observed only in MDR strains (28). In three separate studies performed in other parts of the world on the presence of methicillin and biocides resistance genes, a higher frequency of the qacA/B gene was reported compared to the SMR gene (29-31). The high frequency of these genes in the clinical samples, especially those collected by nasal and urine swabs and blood cultures, indicates the emergence of staphylococcal infections resistant to antibiotics, antiseptics, and disinfectants. In Shi et al. in Hong Kong, the highest frequency of antibiotic resistance genes in gram-positive bacteria was related to the qacE/B gene. The frequency of SMR and qacE genes in coagulase-negative staphylococcus isolates was determined to be 12% (32). In Ignak et al. in Turkey, 71% of Staphylococcus isolates had resistance genes to antiseptics. Still, none of the Enterococcus isolates were reported to have these genes. The frequency of qacA/B and SMR genes in coagulase-negative staphylococci and staphylococcus aureus isolates was 62.5%, 17.5%, and 10.3%, 13.8%, respectively (33). In another study by Mahzounieh et al. , 40% of Acinetobacter baumannii isolates carried the qacE gene, and 80% carried the qacΔE1 gene (34). Moreover, Babaei et al. in Malaysia on the prevalence of resistance genes against QACs among Acinetobacter baumannii isolates demonstrated that 73% of the isolates carried the qacE gene (35). In Qian Y et al. in China between 2010 and 2014, 61.2% of Acinetobacter baumannii isolates carried the qacEΔ1 gene (36). In most studies reviewed in Iran, the highest frequency of resistance genes to antiseptics was qacA/B and SMR in gram-positive isolates and qacE and qacEΔ1 in gram-negative isolates. In these studies, gram-negative isolates containing qacE and qacEΔ1 genes were identified from hospitalized patients in intensive care units and internal diseases and infectious diseases wards from trachea and urine samples. The special conditions of the intensive care units and the use of invasive tools such as urinary catheters in patients were the main causes of infection with resistant organisms. Furthermore, diversity in infection control measures like the type, amount, and concentration of disinfectants used in different hospital departments were other reasons for resistance to antiseptics in medical environments. The results indicated the significant presence of resistance genes to antiseptics in gram-positive and gram-negative isolates in medical centers. Identifying these isolates is particularly important considering the important role of resistant microorganisms in causing various infections and their contribution to increased mortality, especially in patients hospitalized in intensive care units.
5.1. Conclusions
The highest frequency of resistance genes to antiseptics was in qacA/B and SMR for gram-positive isolates and qacE and qacEΔ1 for gram-negative isolates. Considering the ability of these genes to circulate between strains, it is necessary to prevent the spread of antibiotic-resistant strains by applying infection control measures. In addition, identifying genes resistant to antibiotics and antiseptics can also be beneficial in controlling antibiotic-resistant strains. The use of biocidal compounds in the exact concentrations recommended by the manufacturers is also necessary to prevent pollution and reduce the resistance of pathogens to antiseptics.