1. Background
Increased antibiotic resistance makes the need for drug replacement more significant than ever (1). Escherichia coli is one of the leading causes of gastroenteritis (2). E. coli O157:H7 is the most critical serotype among enterohemorrhagic strains of E. coli, playing a role in causing foodborne diseases and being a major contributor to intestinal infections in humans (3). This bacterium leads to severe bloody diarrhea and high-risk diseases like hemolytic uremic syndrome (HUS), hemorrhagic colitis (HC), and thrombotic thrombocytopenic purpura (TTP) (4). Escherichia coli O157:H7 is primarily transmitted through the ingestion of contaminated food or water. It can originate from various sources, including undercooked ground beef, unpasteurized dairy products, raw fruits and vegetables, and contaminated water. Cross-contamination during food handling and preparation, inadequate hygiene practices, and improper sanitation can contribute to the spread of E. coli O157:H7 (5). Escherichia coli strains producing Shiga toxin (STEC) can survive for extended periods in foods such as sausages and mayonnaise (6). Another characteristic of this bacterium is its low infectious dose, which is less than 100. As a result, all enterohemorrhagic E. coli strains found in animals are considered a risk to humans. However, serotype O157 can cause disease in humans and does not pose a risk to adult animals (7). The virulence factors of E. coli O157:H7, including toxins, adhesins, hemolysin, lipopolysaccharide, and flagellum, significantly contribute to the low infectious dose of this bacterium (8). The most critical toxins produced by this bacterium are stx-1 and stx-2 (9). Additionally, Shiga toxin and lipopolysaccharide are the primary factors responsible for causing hemolytic uremia syndrome (10). In contrast to the lower prevalence of diarrheal diseases in developed countries, bacterial diarrhea remains one of the most significant infectious diseases in developing countries (11). Oxadiazoles belong to the group of heterocyclic compounds known for their antibacterial properties, primarily due to their ability to disrupt the integrity of bacterial cell membranes. Oxadiazole derivatives can interact with bacterial cell membranes, leading to destabilization and increased permeability. This disruption results in cell death as ions and proteins leak out of the bacterial cell (12). Another mechanism of action involves the inhibition of essential bacterial enzymes necessary for survival and replication. Oxadiazole derivatives can target specific enzymes involved in critical metabolic pathways of bacteria, such as DNA synthesis, protein synthesis, or cell wall synthesis. By inhibiting these enzymes, oxadiazole derivatives disrupt crucial cellular processes, impeding bacterial growth and viability (13).
Additionally, oxadiazole derivatives may exert antibacterial effects by interfering with bacterial DNA replication and transcription. Enzymes like DNA gyrase and topoisomerase are essential for DNA replication and supercoiling. Oxadiazole derivatives can induce DNA damage and disrupt the accurate replication and transcription of bacterial genetic material by inhibiting these enzymes (14). Some 1, 3, and 4-oxadiazole derivatives have demonstrated antimicrobial properties (15).
1,3,4-Oxadiazoles have the capability to inhibit both Gram-positive bacteria like Staphylococcus aureus, Bacillus subtilis, Staphylococcus epidermidis, and Gram-negative bacteria including E. coli and Pseudomonas aeruginosa (16). Molecular docking is a widely utilized research technique as it enables the prediction and analysis of interactions between small molecules (ligands) and target biomolecules, such as proteins or nucleic acids. This method offers valuable insights into binding affinity, interaction modes, and potential biological activities of ligands when they interact with their target biomolecules.
Researchers can identify potential drug candidates by computationally simulating docking processes, screening extensive compound libraries, and evaluating binding energies and structural compatibility with the targets. Molecular docking plays a critical role in drug discovery and design by enabling researchers to optimize and prioritize compounds for further experimental validation. This approach accelerates the development of novel therapeutics and provides a cost-effective means of identifying potential leads.
2. Objectives
This study had two primary objectives: first, to conduct an in vitro evaluation of the antibacterial effects of certain three-component derivatives of 1,3 and 4-oxadiazole against E. coli O157:H7, and second, to perform an in silico investigation of these compounds regarding their interactions with stx-1 and stx-2 toxins.
3. Methods
The experimental research was carried out in the microbiology laboratory of Islamic Azad University, Tehran branch, in the year 2022. All materials, solvents, and culture media (Nutrient agar/broth) were procured from Merck Co., Germany. The bacterial strain used in the study was obtained from the Iranian Research Organization for Science and Technology (IROST), specifically E. coli O157:H7 ATCC 43890.
3.1. In vitro
All three-component derivatives of 1, 3, and 4-oxadiazole from our previous study were resynthesized (17). To assess their antibacterial activity, the new compounds were solubilized in dimethylsulfoxide (DMSO) solvent at a concentration of 1 mg/mL. Additionally, pure powder of the antibiotic ciprofloxacin (Cip), manufactured by Sigma Company, was prepared and used as a control at a concentration of 1 mg/mL, following CLSI guidelines (18).
The antibacterial activity of the synthesized compounds was investigated using the agar well diffusion method, minimum inhibitory concentration (MIC), and minimum bactericidal concentration (MBC) methods against E. coli O157:H7. Nutrient agar/broth culture medium was prepared according to the brochure instructions. The bacterial sample, obtained from lyophilization, was prepared for testing and evenly spread on the culture plate. Wells of the required size (5 mm) were created on the culture medium using a sterilized Pasteur pipette, and these wells were then filled with solutions prepared from oxadiazole derivatives and the control sample.
The plates were incubated at 37°C for 24 hours, and after this period, they were examined to determine the presence of halos. Subsequently, the inhibitory effect was determined using the tube dilution method to assess the antibacterial effect of the derivatives. Successive dilutions of each derivative (ranging from 15.625 to 1000 µg/mL) were prepared in a nutrient broth culture medium. The highest dilution that prevented bacterial growth (resulting in no turbidity) after 24 hours at 37°C was considered the MIC. Samples with no bacterial growth were then cultured on a nutrient agar medium and the highest dilution that still caused no growth was reported as the MBC (19).
3.2. In silico
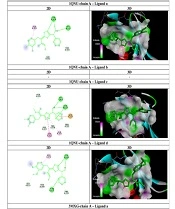
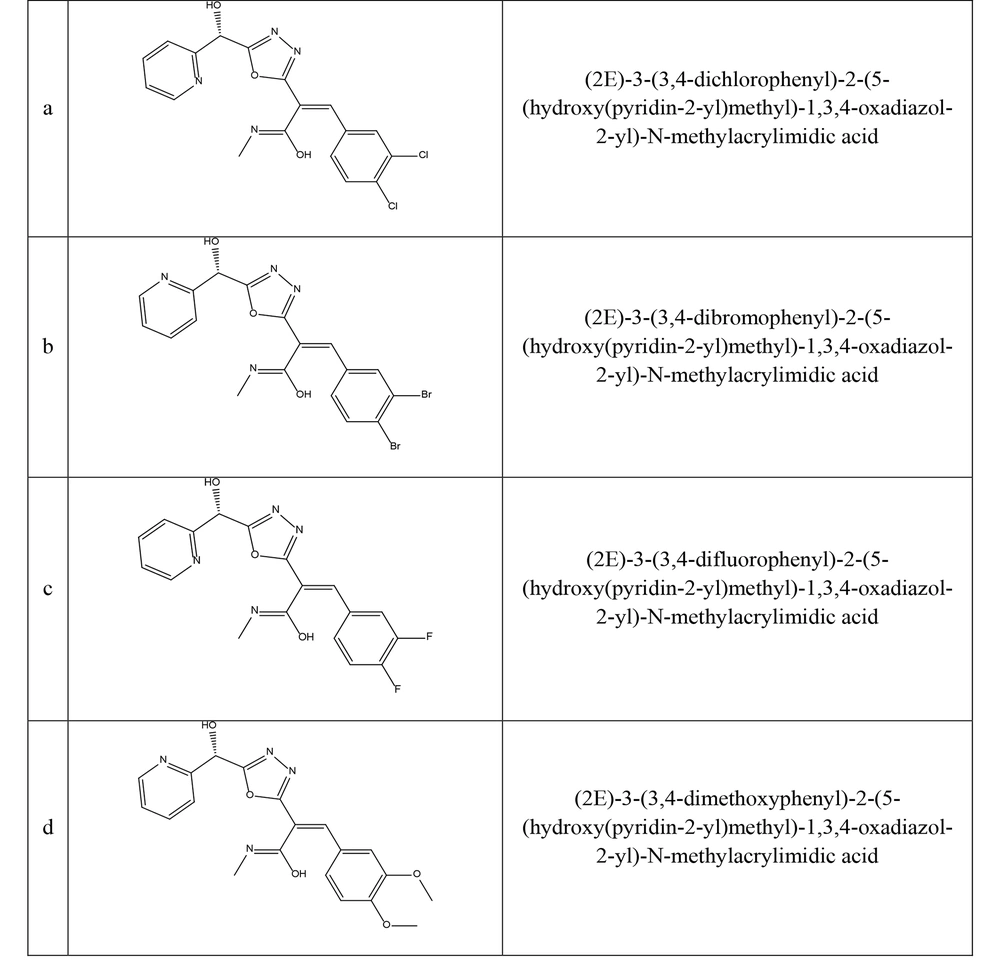
Two-dimensional derivative structures were drawn using ChemDraw 12.0.2.1076 software, and Chem3Dpro 12.0.2.1076 software was employed to standardize the structure with the MM2 method (Figure 1) (20). The crystallized structures of stx-1 (PDB ID: 1QNU-chain A) and stx-2 (PDB ID: 3MXG-chain A) of E. coli O157:H7 were obtained from the Protein Data Bank (PDB) with a resolution of 3°A angstrom. Gastiger charges and polar hydrogens were added using AutoDockTools-1.5.6 software. All water molecules and ligands were removed from the main chains of each structure (Grid box dimensions for 1QNU = 40 × 40 × 40 and 3MXG = 40 × 40 × 40 with a spacing of 3°A).
The docking process of derivatives to the stx-1 and stx-2 binding sites was carried out using AutoDockVina software and Discovery Studio 4.5. The interactions between ligands and binding sites were analyzed using Discovery Studio 4.5 Client software (21).
4. Results
4.1. In vitro
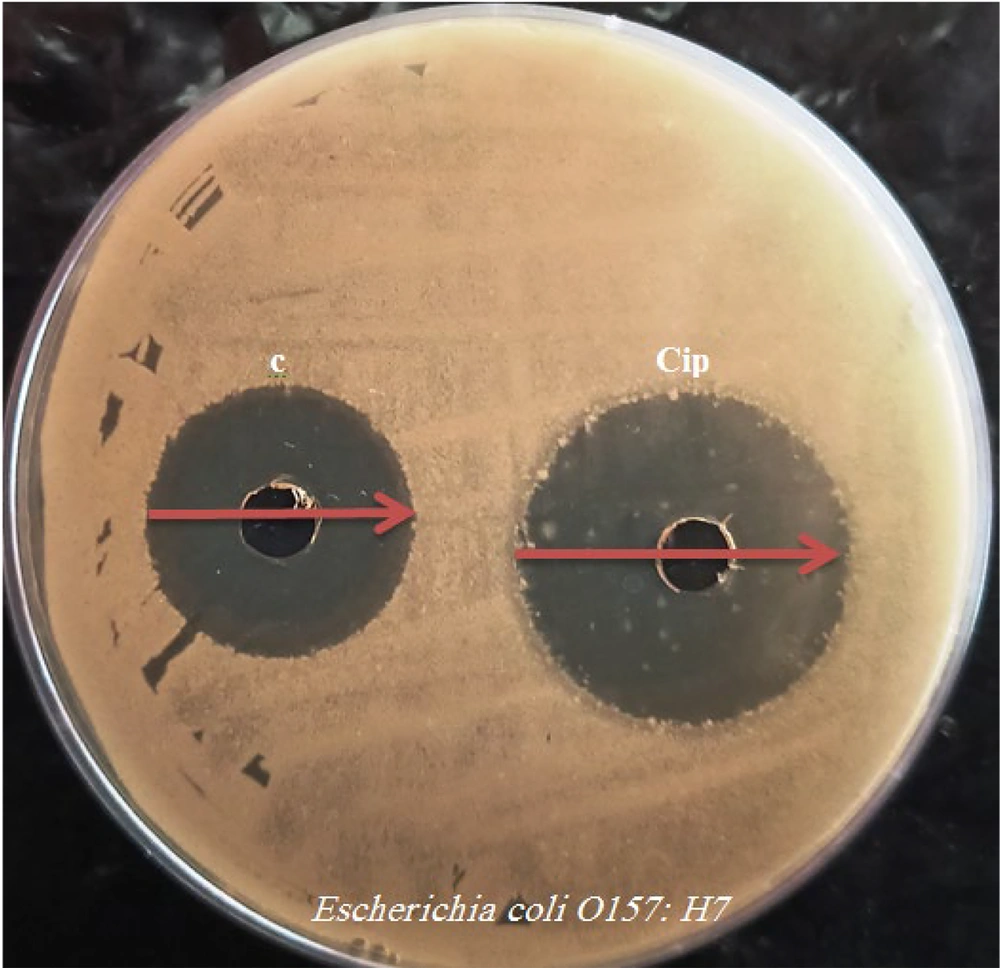
The antibacterial activity of the synthesized 1, 3, and 4-oxadiazole derivatives (a-d) was evaluated at a concentration of 1 mg/mL. The diameters of the inhibition zones (IZ) for each compound are presented in Table 1. Compounds a, c, and d, which contain chlorophenyl, fluorophenyl, and methoxyphenyl groups, exhibited significant antibacterial activity (Figure 2). Compound b, on the other hand, did not display notable antibacterial activity. Among the synthesized compounds, compound c stood out with an IZ of 36 ± 0.5mm, MIC of 62.50 µg/mL, and MBC of 125 µg/mL against E. coli O157: H7. Compound c showed effects similar to the control sample (Figure 2).
| Compounds | Inhibition Zone, mm | Minimum Inhibitory Concentration, µg/mL | Minimum Bactericidal Concentration, µg/mL |
|---|---|---|---|
| a | 25 ± 0.5 | 500 | 1000 |
| b | 12 ± 0.5 | 1000 | 1000 |
| c | 36 ± 0.5 | 62.50 | 125 |
| d | 26 ± 0.5 | 500 | 1000 |
| Cip | 42 ± 0.5 | 31.25 | 62.50 |
Antibacterial Activity of 1, 3, 4-Oxadiazol Derivatives
4.2. In silico
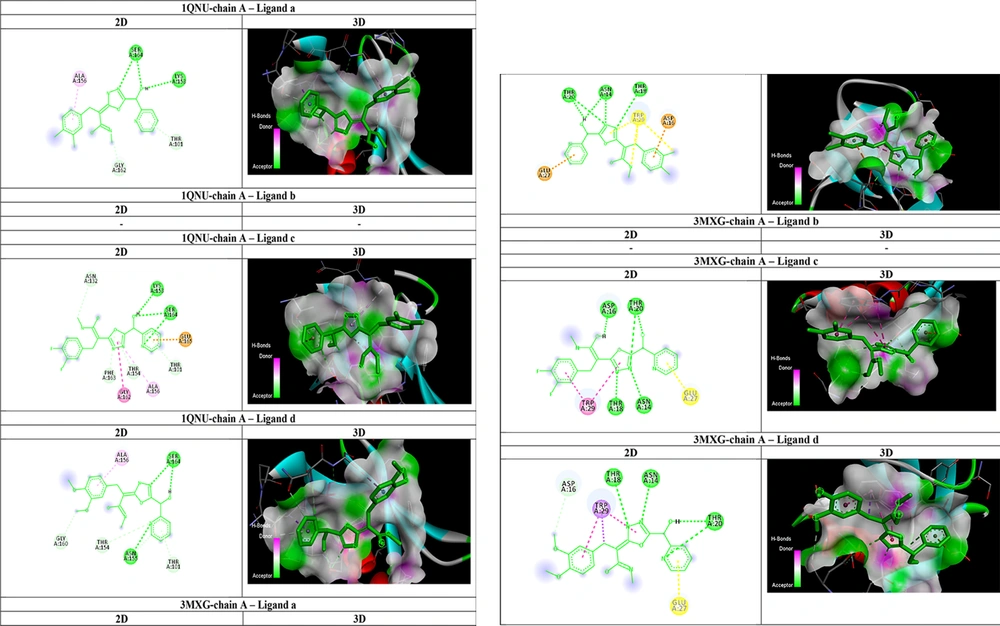
As shown in Table 2, all affinities were calculated, and the ligand with the best affinity (lowest ΔGbind) for each receptor was selected for further AutoDock interactions. The interactions of ligands within the active site of 1QNU-chain A and 3MXG-chain A are detailed in Table 2 and Figure 3. Ligand c ((2E)-3-(3,4-difluorophenyl)-2-(5-(hydroxy(pyridin-2-yl)met hyl)-1,3,4-oxadiazol-2-yl)-N-methylacrylimidic acid) formed two strong hydrogen bonds with the amino acids lysine 153 and serine 164, as well as three robust carbon-hydrogen bonds with threonine 101, threonine 154, and phenylalanine 163, within the stx-1 receptor. The docking results indicated that ligand c adopted a favorable conformation and established stronger bonds, resulting in greater inhibition of the stx-2 receptor compared to other receptors. For stx-2 receptor interaction, ligand c formed four strong hydrogen bonds with the amino acids asparagine: 16, threonine 20, threonine 18, and arginine 14, along with one robust carbon-hydrogen bond with tryptophan 29. Ligand c, which features a fluorophenyl group, exhibited valuable inhibitory potential and could serve as a promising candidate for the development of alternative drug structures in the treatment of E. coli O157: H7 infections in future research.
| Ligand and Receptor | Affinity, kCal/mol | Hydrogen Bonds | Carbon-Hydrogen Bonds | Pi-Alkyl, Pi-Anion, and Pi-Amide Bonds |
|---|---|---|---|---|
| a | ||||
| 1QNU-chain A (stx-1) | -6.5 | Serine 164, Lysine 153 | Glycine 162, Threonine 101 | Alanine 156 |
| 3MXG-chain A (stx-2) | -6.9 | Threonine 20, Threonine 18, Asparagine 14 | - | Asparagine 16, Glutamine 27 |
| b | ||||
| 1QNU-chain A (stx-1) | -6.5 | - | - | - |
| 3MXG-chain A (stx-2) | -7.0 | - | - | - |
| c | ||||
| 1QNU-chain A (stx-1) | -7.4 | Lysine 153, Serine 164 | Threonine 101, Threonine 154, Phenylalanine 163 | Glutamine 165, Alanine 156, Glycine:162 |
| 3MXG-chain A (stx-2) | -8.9 | Asparagine 16, Threonine 20, Threonine 18, Arginine 14 | Tryptophan: 29 | - |
| d | ||||
| 1QNU-chain A (stx-1) | -6.2 | Serine 164, Asparagine 155 | Threonine 154, Threonine 101, Glycine 160 | Alanine 156 |
| 3MXG-chain A (stx-2) | -6.7 | Threonine 18, Threonine 20, Arginine 14 | Asparagine 16 | Tryptophan 29 |
AutoDockVina Results of 1, 3, 4-Oxadiazole Derivatives as an Inhibitor of stx-1 and stx-2 of Escherichia coli O157: H7
5. Discussion
The global prevalence and pathogenicity of EHEC, coupled with the fact that conventional antibiotics can enhance their pathogenicity, emphasize the importance of prevention (22). Beta-lactam antibiotics, including penicillins, cephalosporins, monobactams, and carbapenems, represent some of the most commonly used antibacterial drugs. Nowadays, many bacterial strains have developed resistance to these antibiotics either through genetic mutations or by producing beta-lactamase enzymes (23). Extended-spectrum beta-lactamases (ESBLs) constitute a group of enzymes that confer bacterial resistance to penicillins, cephalosporins (particularly third-generation cephalosporins), and monobactams. Carbapenems like imipenem and meropenem are frequently employed to combat such strains, but their overuse has led to the gradual emergence of drug resistance. Among Gram-negative bacteria, species within the genera Escherichia, Klebsiella, Proteus, Enterobacter, Cerasia, Citrobacter, Salmonella, Pseudomonas, Burkholderia, and Acinetobacter are among the most significant producers of this type of beta-lactamase (24).
Shiga toxin-producing E. coli was first identified as the causative agent of diarrhea in Canada in the late 1970s. Certain strains of E. coli, particularly E. coli O157:H7 and some other enterohemorrhagic E. coli (EHEC) strains, produce stx-1 (Shiga toxin 1) and stx-2 (Shiga toxin 2). These toxins are encoded by bacteriophages, viruses that infect bacteria. The production of stx-1 and stx-2 involves a two-step process: lysogenic conversion and toxin expression (25).
Vero toxins are classified into two groups: stx-1 and stx-2. Verotoxin hinders protein synthesis by targeting ribosomal RNA. Escherichia coli strains that produce these toxins can lead to dangerous syndromes, including hemorrhagic colitis followed by kidney failure, especially in children, as well as thrombotic thrombocytopenic purpura in humans (26).
Oxadiazoles exhibit a wide range of biological activities (27). For example, Shah et al. investigated the antibacterial effects of new S-substituted derivatives of 5-[3-(1H-indol-3-yl) propyl]-1,3,4-oxadiazol-2-ylhydrosulfide and found that most of the compounds demonstrated favorable antibacterial activity against Bacillus subtilis and E. coli (28). In our research, derivative c ((2E)-3-(3,4-difluorophenyl)-2-(5-(hydroxy(pyridin-2-yl)met hyl)-1,3,4-oxadiazol-2-yl)-N-methylacrylimidic acid) exhibited strong antibacterial activity against E. coli O157: H7. The presence of the fluorophenyl functional group in the synthesized structure may contribute to its antibacterial effect. In line with this, Kariuki et al. synthesized fluorophenyl structures and demonstrated that these compounds inhibited the growth of both Listeria monocytogenes and E. coli, with activity levels comparable to those of ampicillin and vancomycin (29).
Our study assessed compounds a-d as ligands for inhibiting the stx-1 and stx-2 receptors. The results indicated that compound c had a higher ability to inhibit stx-2 compared to the others due to its strong hydrogen bonds. Compounds a-d were evaluated as ligands for inhibiting the stx-1 and stx-2 receptors, and compound c showed a greater ability to inhibit stx-2 than stx-1, primarily because of its strong hydrogen bonds. In a related study, Luo et al. conducted antibacterial evaluation and molecular docking studies of 1,3,4-Oxadiazole-2(3H)-Thione-norfloxacin hybrids as potent antibacterial agents and reported an enhanced receptor-hybrid affinity towards DNA topoisomerase IV, with a minimum binding energy of -9.4 to -10.0 kcal/mol. Among all the synthesized hybrids, 3e could be a potential candidate for further antibiotic development (30). Computer data has indicated that oxadiazole compounds with high levels of stx-2 inhibition could potentially mitigate the pathogenicity of E. coli O157: H7. This possibility awaits confirmation through In vivo tests. Overall, the current theoretical data suggest that the compound (2E)-3-(3,4-difluorophenyl)-2-(5-(hydroxy(pyridin-2-yl)meth yl)-1,3,4-oxadiazol-2-yl)-N-methylacrylimidic acid may have the capacity to inhibit the E. coli O157: H7 pathogen. The results imply that the potential anti-E. coli O157: H7 effect of 1, 3, and 4-oxadiazole compounds against the factors involved in the pathogenicity of this bacterium may be attributed to the inhibition of various receptors, including stx-2. The findings of this research could play a pivotal role in the development of therapeutic drugs for E. coli O157: H7.