1. Background
Breast cancer is one of the main causes of death in women. Studies show that the mortality rate of breast cancer is higher in less-developed areas. It is estimated that in 2008, about 8 million deaths occurred due to breast cancer, which is likely to reach 11 million in 2030 (1). There are various treatments for cancer, such as radiation therapy and chemotherapy (2). The anti-cancer drugs, has two major problems, which are drug resistance and side effects (3). One of the ways to reduce the problems of using anti-cancer drugs is to use combination treatment, and in this field, materials of natural origin are one of the solutions considered by researchers (4).
European Medicines Agency (EMA) has approved everolimus as an effective drug for the treatment of advanced breast cancers with positive and negative hormone receptors (5). Everolimus has inhibitory effects on the aggressiveness of breast cancer cells and causes the cell cycle to stop. This activity of everolimus is due to the PI3K/AKT/mTOR signaling pathway inhibition, which subsequently increases the expression level of caspases 3 and 8 in breast cancer cells (6). Also, research has shown that everolimus inhibits complex 1 (mTORC1) and also induces autophagy, and arrests the G1 cell cycle. Everolimus can stimulate the destruction of cyclin D1 by autophagy in breast cancer cells. Everolimus stimulates autophagy and reduces the level of Cyclin D1 is destroyed in breast cancer tissue; everolimus-induced autophagy in breast epithelial cells destroys cyclin D1 and, as a result, stops the G1 cell cycle (7).
Bee venom is a complex mixture of various compounds, including peptides, enzymes, and non-peptide components. Melittin (40 - 50%) of bee venom has been reserved for itself. Melittin protein, which is a part of bee venom and a toxic peptide soluble in water, has antiparasitic, antibacterial, antiviral, and anti-inflammatory activities. Melittin has a wide range of antitumor and cytotoxic effects on different types of tumor cells. it is effective in the treatment of human breast, prostate, bladder, lung, liver, and stomach cancers (8, 9). Melittin rapidly binds to the phospholipid cell membrane and moves along the membrane, causing structural defects and creating pores in the cell membrane. In addition, when it reaches the intracellular environment, it can act similarly on the internal membrane of the organelles, and by creating oxidative stress, it causes biochemical changes that eventually lead to cell death on A375 and MCF7 cell lines. It inhibits PI3K/Akt/mTOR signaling pathway in breast cancer cells (10).
Autophagy has two functions in tumor cells: Tumor suppressor and tumor inducer. In fact, in the early stages of the tumor, it causes suppression, and in the advanced stages, it causes the survival of tumor cells and in this way, it plays a dual role in the growth of tumors. Inhibition of autophagy can be a promising approach for advanced cancers, and also autophagy can be a promising target for drug production in cancer (11). Also, autophagy is a conserved homeostatic process that acts as a cellular protective mechanism and activates in response to anti-neoplastic agents. Inhibition of autophagy can increase the death of tumor cells with various anticancer treatments, which represents an attractive approach to control drug resistance mechanisms (12).
Autophagy can be induced by melittin in hepatocellular carcinoma cells. Inhibition of autophagy by chloroquine (CQ) increased the antitumor effect of melittin, but induction of autophagy by RAPA decreased the effect of melittin in hepatocellular carcinoma cells (13).
2. Objectives
Considering that both melittin and everolimus substances have anti breast cancer activities and the combined effect of these two substances has not been investigated; therefore, we decided to study the simultaneous effect of these two drugs on the autophagy in human breast cancer cell lines.
3. Methods
3.1. Cell Culture and Treatment
Human breast cancer cells were purchased from Pasteur Institut of Iran. They were seeded in RPMI medium, 10% Fetal Bovine Serum (FBS), and without antibiotics. The cell line was kept in the incubator at a temperature of 37°C, 90% humidity, and 5% carbon dioxide (14). Cells were treated with melittin (0.01, 0.03, 0.06, 0.12, 0.25, 0.5, 1, 2 µM) and everlimus (0.01, 0.03, 0.06, 0.12, 0.25, 0.5, 1, 2 µM) for 24 and 48 and 72 and 96 hr for MTT assay. For other tests, concentrations of IC50 and treatment duration of 24 hr were used.
3.2. MTT Test to Measure Cell Survival
For this test, a suitable number of cells were seeded in wells of a 96-well plate and incubated overnight. Then, the medium containing the desired concentration of each drug was added to each of the wells (first, the initial test was performed and the IC50 of each agent was calculated alone. Then the cells were again treated with two concentrations higher and lower than IC50), and after the specified time, MTT solution (usually 50 µL) was added to each wells (15). The cells were kept in the incubator for 3 hr in the dark. DMSO was added to each of the wells. After that the absorbance was measured at 570 nm using a microplate reader. Control Cells were treated with culture medium without serum (the solvent for melittin and everlimus). The percentage of cell viability was calculated from the following Equation:
(A570 of treated cells/A570 of untreated cells) × 100
In the next step, the treatment was performed by combining two agents at the same time in two concentrations higher and two concentrations lower than IC50 of melittin and everlimus. In the last step, the data was assessed by Compusyn software and the combination index (CI) and dose reduction index (DRI) parameters were calculated (15).
3.3. Autophagy Assay
Autophagy is the process of packing cytoplasmic constituents in disintegrating organelles, and this process is the formation and development of acidic vesicular organelles. Acridine orange accumulates in acidic organelles in the cell in a pH-dependent manner. This dye emits green fluorescence at neutral pH, but is protonated in an acidic environment, trapped inside the organelle, and forms aggregates that emit red fluorescence (16). The cells were cultured in a 24-well plate, attached to the bed and treated. After that, the wells were stained by acridine orange solution with 1 μg/mL dissolved in PBS for 15 min. The stained samples were examined by fluorescence microscope. These three load experiments were repeated at three different times and in each amount, one hundred cells were randomly selected in the microscopic fields. The percentage of autophagic cells was calculated using the following formula Equation:
(Total number of cells/number of cells with acidic vesicular organelles) × 100
3.4. Gene Expression Analysis
The expression levels of Atg-7, Beclin-1, and lC-3 were analyzed by real-time PCR assay. The cells were treated for 24 hr, RNA was extracted by by DENAzist total RNA isolation kit (Tehran, Iran). A nanodrop spectrophotometer and electrophoresis methods were used for verified the quantity and quality of the RNAs, respectively. The complementary DNA (cDNA) synthesis was carried out using Vivantis Technologies cDNA synthesis kit (Selangor, Malaysia) according to the manufacturer’s protocol. GAPDH served as a housekeeping gene. Real-time PCR was done using Takara Bio Inc. SYBR Premix Ex Taq Technology (Shiga, Japan). The expression of genes was estimated based on the comparative Ct (2-ΔΔct) method. The primer sequences were designed using GeneRunner software and listed in Table 1 (17).
| Genes | Primer |
|---|---|
| Atg-7 | |
| Forward | 5-ATTGCTGCATCAAGAAACCC-3 |
| Reverse | 5-GATGGAGAGCTCCTCAGCA-3 |
| Beclin-1 | |
| Forward | 5-GCCGAAGACTGAAGGTCA-3 |
| Reverse | 5-GTCTGGGCATAACGCATC-3 |
| LC-3 | |
| Forward | 5-GATGTCCGACTTATTCGAGAGC-3 |
| Reverse | 5-TTGAGCTGTAAGCGCCTTCTA-3 |
| GAPDH | |
| Forward | 5-TCCCTGAGCTGAACGGGAAG-3 |
| Reverse | 5-GGAGGAGTGGGTGTCGCTGT-3 |
Primer Sequences for RT-PCR Analysis
3.5. Statistical Analysis
The tests were done in triplicate and data were expressed as means ± Standard Error of Mean. Statistical analysis was done by Tukey’s test one-way analysis of variance and differences were considered significant when P < 0.05.
4. Results
4.1. Viability Assay
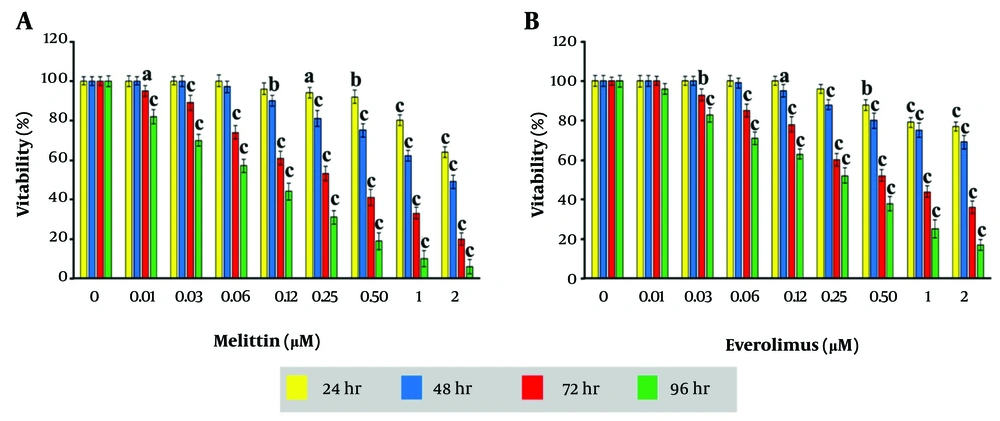
The effect of melittin and everolimus on cell viability is shown in Figure 1A and B. Both melittin and everolimus decreased cell viability in a dose- and time-dependent manner, after 24, 48, 72, and 96 hr. IC50 values for melittin were 4.25, 1.62, 0.32 and 0.1 µg/mL and for everolimus were 5.38, 2.81, 0.67, and 0.26 µg/mL for 24, 48, 72, and 96 hr, respectively.
The CI values in all combinations groups were less than 1, indicating a synergistic effect. The DRI values were more than 1 indicating a concentration decrease for a given therapeutic effect in both of them (Table 2).
| Co-treatments Groups | Fa | CI | DRI Melittin | DRI Everolimus |
|---|---|---|---|---|
| 1 | 0.34 ± 0.05 | 0.96 | 2 | 2.11 |
| 2 | 0.55 ± 0.04 | 0.81 | 2.47 | 2.43 |
| 3 | 0.68 ± 0.09 | 0.92 | 2.22 | 2.09 |
| 4 | 0.82 ± 0.07 | 0.85 | 2.5 | 2.2 |
| 5 | 0.89 ± 0.05 | 0.95 | 2.29 | 1.92 |
Fraction Affected, Combination Index and Dose Reduction Index Values for Melittin and Everolimus Combination a
4.2. Autophagy Assay
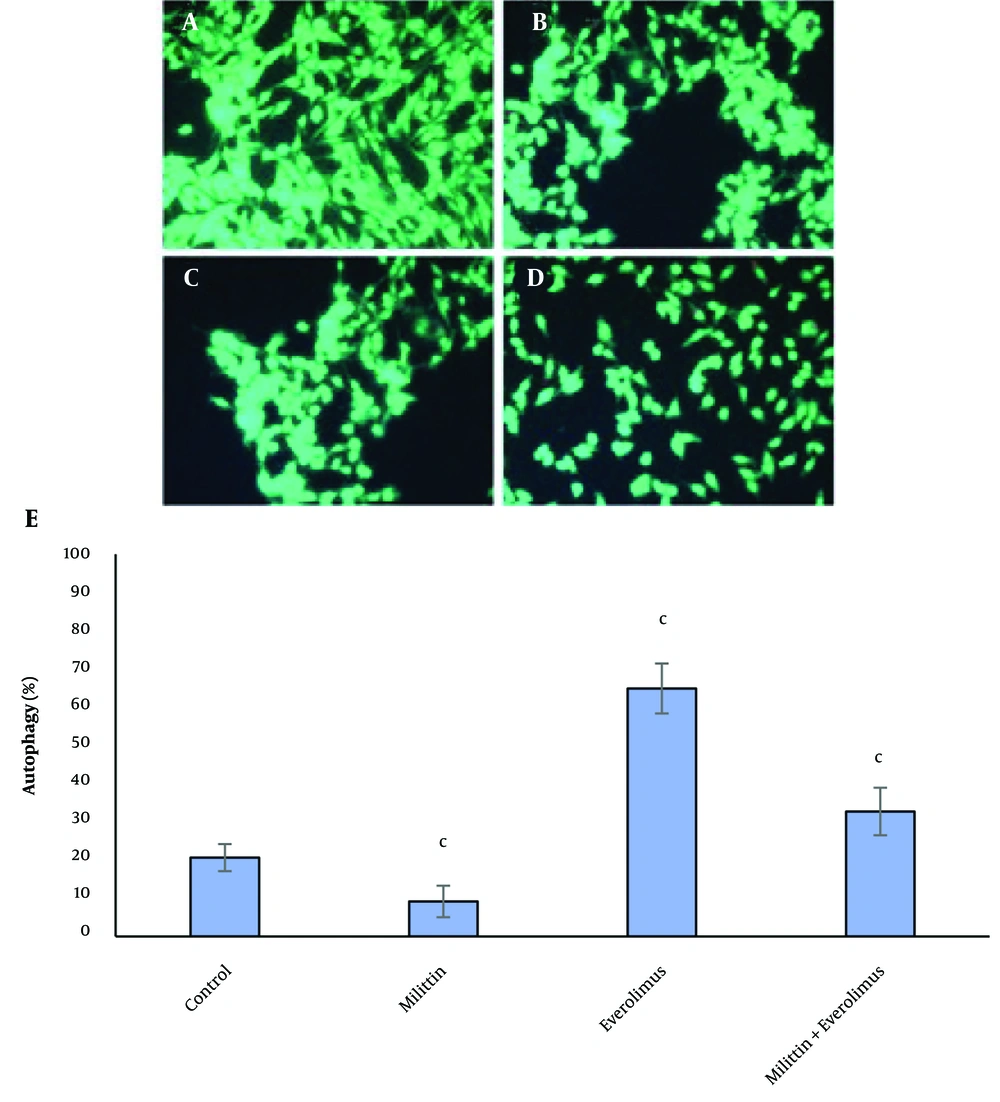
A little cytoplasmic staining was seen in the control cells and cells treated with melittin, however, the everolimus-treated tumor cells showed abundant autophagy. Autophagy was decreased significantly in the combination group compared with everolimus-treated group (Figure 2).
The effects of melittin and everolimus on autophagy in breast cancer cells. A, control cells; B, in the presence of IC50 concentration of melittin, C, in the presence of IC50 concentration of everolimus; D, in the presence of IC50 concentration melittin and of IC50 concentration everolimus; and E, columns mean percentage of autophagic cells from three independent experiments. Red dots indicate autophagic vesicles. The data are expressed as the percentage of the control cells as the means± SEM. c, P < 0.01 compared with control.
4.3. Real-time PCR Assay
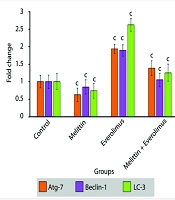
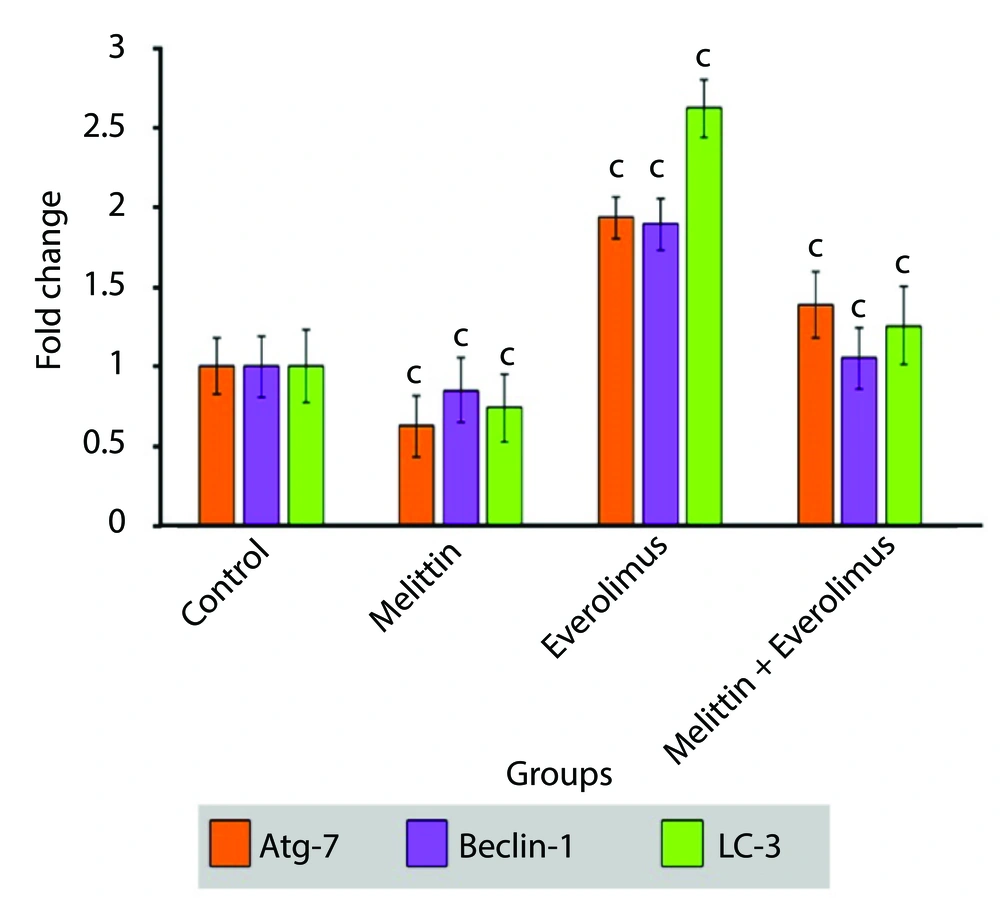
Autophagy-related genes expression was tested by real-time PCR. Everolimus increased the expression of three autophagy marker genes (Atg-7, LC-3, Beclin-1). Melittin decreased their expression. Their expression was decreased significantly in the combination group compared with everolimus-treated group (Figure 3).
5. Discussion
In this study, melittin and/or everolimus anti breast cancer effect was evaluated. The data of the present study showed that after all four periods of treatment with melittin, cell viability decreased gradually with increasing concentration. After 24, melittin decreased the cell viability significantly in the concentrations of 0.25, 0.5, 1 and 2 μg/mL compared to the control group. After 48 hr, the decrease in survival was significant in concentrations of 0.12, 0.25 ,0.5, 1, 2, μg/mL compared to the control group. After 72 and 96 hr of treatment, the effect of melittin on reducing survival was significant in all groups. IC50 values for melittin were 4.25, 1.62, 0.32 and 0.1 µg/mL for 24, 48, 72, and 96 hr, respectively.
In a study close to our study, in relation to breast cancer (MDA-231 cell line), it was shown that after 24 hr treatment, melittin’s IC50 was measured 3.8 μg/mL (18). This value of IC50 is close to our study. Also another study measured the effect of melittin on the viability of human hepatocellular carcinoma cells (HepG2). The IC50 values for melittin in HepG2 cells were 5.94, 5.65, and 4.87 µg/mL for 24, 48, and 72 hr, respectively (19). IC50 values obtained in our study were slightly lower than this study, which is probably due to the higher sensitivity of breast cancer cells to melittin.
Previous study showed that melittin inhibited invasion and migration of breast cancer cells via mTOR pathway inhibition (20). Melittin shows significant antitumor activity against various human cancer cell lines, especially it can suppress cell viability and promote apoptosis in HCC cells. It is affect the expression of proteins in the mitochondrial apoptosis pathway. It can induce autophagy in breast cancer cells (18, 21). Melittin is highly effective candidate for cancer immunotherapy (21). Study of melittin effects on gastric and colorectal cancer (AGS, COLO205 and HCT-15 cell lines) showed that melittin had a dose-dependent effect at 4 h of treatment, with complete cell death at the highest dose of 20 μg/mL. Interestingly, melittin induced cellular changes within seconds when observed at shorter time points. After 1 min of melittin treatment, membrane changes were observed and intracellular substances were observed to be removed from the cells (22).
After 24 hr of treatment with everolimus, the decrease in survival was significant at concentrations of 0.25,0.5,1,2 µ/mL compared to the control group. After 48 hr, the decrease in survival in concentrations of 0.12, 0.25, 0.5, 1, 2 μg/mL was significant compared to the control group. Also, after 72 and 96 hr, the decrease in survival in concentrations of 0.03, 0.06, 0.012, 0.25, 0.5, 1, 2 μg/mL was significant compared to the control group. IC50 values for everolimus were 5.38, 2.81, 0.67, and 0.26 µg/mL for 24, 48, 72, and 96 hr, respectively.
Everolimus has been used in combination with other drugs such as capecitabine in HER2 metastatic breast cancer. which had encouraging results for survival and clinical benefit in patients with metastatic breast cancer (23). Also, everolimus decrase the growth of cultured breast cancer cell lines by inhibition of the mTOR pathway, which is similar to melittin (24). Combination of miR-7-5p with everolimus induces apoptosis to determine anticancer therapeutic effect in non-small cell lung cancer treatment (25).
Despite current advancements in cancer medicine, drug resistance is still one of the major barriers to cancer treatment. Thus, it is crucial to provide new solutions to overcome this issue (26). Today, drug combination is the best strategy for treating diseases such as cancer. This strategy reduces the dose and toxicity, and minimize or delay drug resistance. The use of several anticancer drugs is widely used in the treatment of various cancers. Previous study showed that the combination of everolimus and paclitaxel can be used for adriamycin-resistant breast cancer treatment. Everolimus increased the effect of paclitaxel, prevented cell proliferation and induced apoptosis, and stoped the cell cycle in the G1 phase, by reducing the expression of the mTOR signaling pathway proteins (26).
In this study, the CI values were between 0.92 and 0.81 for the combination of melittin and everolimus, which indicated the synergistic effect. The average CI for all experiments was 0.91, which indicated the overall synergistic effect of the combination of melittin and everolimus on breast cancer cells.
The data indicated that autophagy was increased significantly in the treatment with everolimus, and decreased significantly in the treatment with melittin. Finally, in the simultaneous treatment, autophagy was significantly lower than everolimus treated group, but no significant difference was observed with control cells. Also, investigations at the molecular level showed that the mRNA expression of autophagy stimulating genes (Atg-7, beclin-1 and LC-3) was significantly increased by Everolimus and decreased by melittin. These results were in full agreement with the results of fluorescence staining of the cells. Induction of autophagy by everolimus can be one of the main reasons for cell resistance to this drug, and its inhibition at the transcription level by melittin increases the sensitivity of cells to drug toxicity and increases its effectiveness.
5.1. Conclusions
We conclude that, melittin reduces the resistance of human breast cancer cells or increases their sensitivity to everolimus through blocking of autophagy process and consequently more breast cancer cells eliminate