1. Background
Enterococci, the primary opportunistic pathogens implicated in nosocomial infections, are naturally occurring commensal bacteria in the gastrointestinal tracts of humans and animals (1). A significant issue from the past to the present is the deadly opportunistic disease in developing nations, causing a rise in global mortality rates. Diverse microbial agents, including fungi, bacteria, parasites, and viruses, are known to trigger lethal opportunistic infections (2-4). Enterococci usually do not affect healthy individuals, but those with severe underlying health conditions or weak immune systems are at risk. These bacteria are among the most common causes of hospital-acquired infections, leading to bacteremia, endocarditis, and urinary tract infections. Enterococci can enter the environment via sewage systems, and strains resistant to vancomycin are particularly problematic in hospitals, contributing to the prevalence of hospital-acquired infections (5-7). The main reason for their survival in hospitals is their natural resistance to many antibiotics, and they can become resistant to all antibiotics through genetic mutations or acquiring external genetic elements like plasmids and transposons. Vancomycin is often the drug of choice to combat Enterococci, which has developed resistance to multiple drugs. Studying vancomycin-resistant Enterococci (VRE) epidemiology is crucial in environmental contexts. Determining the patterns of antibiotic resistance in VRE strains is essential to prevent their spread (8). The occurrence of VRE differs globally, with significant cases reported in the United States, Britain, Ireland, Saudi Arabia, and Turkey. Conversely, countries like France and Italy report a lower incidence of VRE (9). According to a study by Wada in Nigeria, Enterococcus fecalis was the most, and the pooled prevalence of VRE was estimated at 25.3% (10). In addition, Vancomycin-resistant Enterococci in hospitals on European borders (Netherlands and Germany) were increasingly significant. The biggest difference between the two countries was the studies in Germany showed an increasing trend prevalence in VRE hospitals, and no such trend was observed in studies in the Netherlands (11).
2. Objectives
Previous studies have highlighted the increasing resistance of Enterococcus to vancomycin as a significant concern that calls for epidemiological research. This research analyzes the resistance patterns of vancomycin-resistant Enterococcus strains found in Zahedan.
3. Methods
3.1. Clinical Sampling and Laboratory Identification
In this descriptive-analytical study, among 3000 clinical samples, including Blood culture, Urine, and other fluids, 110 Enterococcus spp. isolates, including E. faecalis and E. faecium, were collected from patients admitted to Ali ibn Abi Talib Hospital in Zahedan, Iran from March 2019 to February 2022. The isolates were cultured in Chocolate and Blood agar Media (Merck, Germany). The causative bacteria were identified based on colony characteristics, Gram stain morphology and biochemical tests (catalase test, growth on bile aesculin agar, tolerance to 6.5% NaCl, arginine dihydrolase test, and fermentation of arabinose, mannitol, raffinose, and sorbitol). The study population included patients of all age groups and both sexes.
3.2. Antibiotic Susceptibility of Enterococcus Species
An antimicrobial susceptibility test using the disc diffusion test (DDT) was carried out for all the confirmed isolates of E. faecalis and E. facume. However, according to the Clinical and Laboratory Standards guidelines, the isolates were categorized as sensitive, resistant, or intermediate to each antibiotic by measuring the respective zone of inhibition. (CLSI-2021) (12). The DDT and antibiotic discs for each pathogen were selected based on CLSI-2021 recommendations as vancomycin (30 μg), ampicillin (10 μg), nitrofurantoin (15 μg) (for urine samples), ciprofloxacin (5 μg), and linezolid (30 μg) (MAST, UK).
3.3. Data Analysis
The data were statistically analyzed using GraphPad Prism version 8 (GraphPad Software, Inc., CA, US) and an appropriate statistical test, i.e., Student’s t-test, Wilcoxon test, chi-square test, or one-way ANOVA.
4. Results
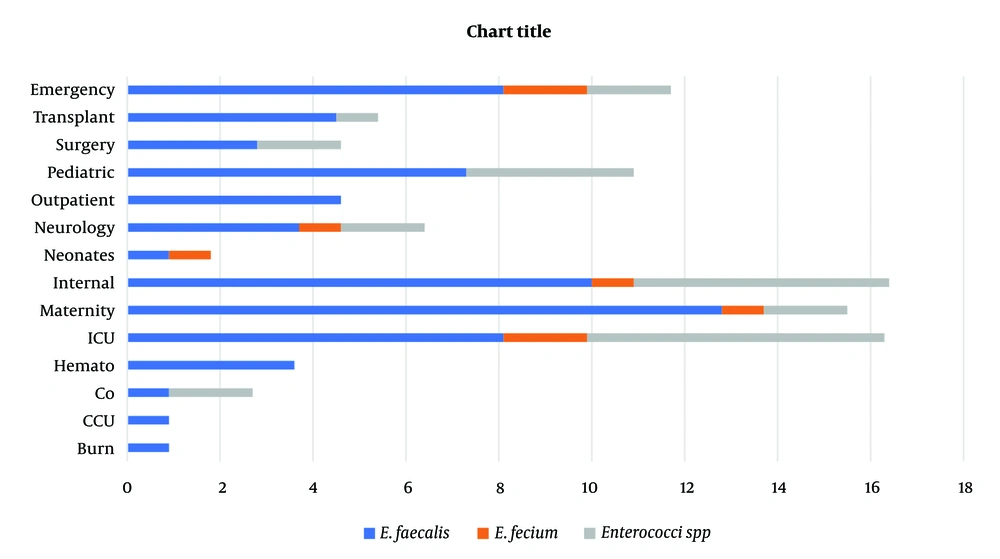
A total of 110 Enterococci were isolated from 3000 culture-positive clinical samples. About 92 urine isolates, 14 blood isolates, one tracheal, two wounds, and one pulmonary isolate were identified as Enterococci (Table 1). All isolates were further speciated as E. faecalis (69.1%), E. faecium (7.3%) and Enterococcus spp. (23.6%). The isolates that were normal flora were not considered.
| Samples | Isolates of Enterococcus faecalis, N = 76 | Isolates of Enterococcus faecium, N = 8 | Isolates of Enterococcus spp., N = 26 |
|---|---|---|---|
| Urine | 70 (92.1) | 5 (62.5) | 17 (65.3) |
| Blood | 3 (3.9) | 3 (37.5) | 8 (30.7) |
| Tracheal | 1 (1.3) | - | -- |
| Wound | 1 (1.3) | - | 1 (3.8) |
| Pulmonary | 1 (1.3) | - | - |
Incidence and Distribution of Enterococcal Isolates in Different Clinical Samples a
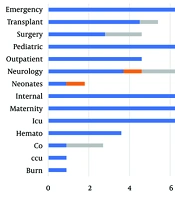
Prevalence of clinical strains in different wards. As shown in Figure 1, isolates from internal (n = 18), ICU (n = 18), and maternity (n = 17) units were the higher isolates (Figure 1).
4.1. Prevalence of Antibiotic Resistance
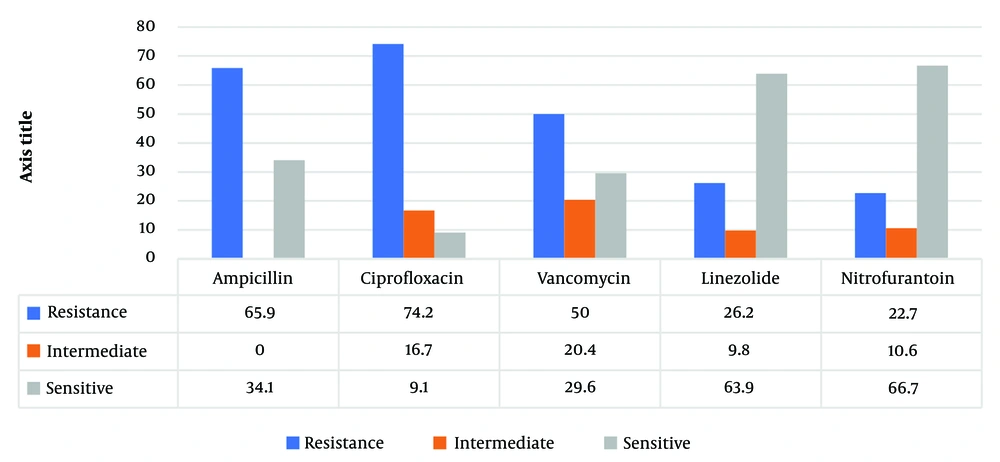
The most frequent resistance was observed for ciprofloxacin and ampicillin, with 74.2% and 65.9% of the isolates, respectively, followed by a 50% resistance rate to vancomycin. On the other hand, resistance to nitrofurantoin and linezolid was comparatively less common, detected in 22.7% and 26.2%, respectively (Figure 2).
5. Discussion
Given the limited treatment options and increasing prevalence of VRE in the world, VRE remains a severe problem in health care. However, this overall increase in variation has been reported in countries. The patterns of antimicrobial resistance and the prevalence of VRE were examined in Zahedan. This investigation indicated a severe problem of antimicrobial resistance among Enterococci in a hospital in Zahedan. In addition, this pathogen was recognized as a significant nosocomial pathogen causing substantial morbidity and mortality globally. Since the studies have shown the rate of VRE in Germany, the UK, and Italy were 11.2%, 8.5%, 12.5%, and 9%, respectively, in the present study, the 50% rate of VRE prevalence was in agreement with earlier reports of high VRE prevalence in other cities. According to a meta-analysis by Emaneini et al., the estimated prevalence of VRE in Iran was 9.4% (13), whereas it was found to be 50% in the present study. The predominant location of Enterococcal infection in this investigation was the urinary tract (84.7%), frequently observed in patients. Septicemia, at 11.2%, was the subsequent most frequent infection after bacteremia. The dominant species of Enterococcus was identified as E. faecalis, accounting for 82% of cases, aligning with the findings of Chakraborty et al. study in India, which reported a prevalence of 80 - 90% for this bacterium in 2015 (11). The bacteria showed high resistance to gentamicin and cephalexin, while resistance to linezolid was the least observed, and 24% of isolates were resistant to vancomycin (8). However, this study noted a 50% resistance rate to vancomycin. In Razaz Rahmati et al., among 203 Enterococcus strains from Tehran hospitals, resistance was 47.3% to ampicillin, 24.6% to vancomycin, and 9.4% to teicoplanin, with a higher sensitivity to linezolid reported (14). The findings for ampicillin resistance and linezolid sensitivity are consistent with these figures. Goudarzi et al. found no resistance to linezolid among 690 Enterococcal isolates (0%) (15), whereas our study revealed a 63.9% sensitivity rate (Figure 2), indicating a notable level of resistance. Abamecha et al. in Ethiopia found that out of 114 isolated Enterococci, 32.4% were resistant to nitrofurantoin, 49.1% to ciprofloxacin, and 36% to ampicillin, showing an increase in resistance compared to previous results (16). Melese et al. reported that 60.7% of VRE strains were resistant to penicillin and 56.5% to amoxicillin, with a relatively low resistance of 13% to linezolid and daptomycin, which is in contrast to the high sensitivity to linezolid observed in the current study (Figure 2) (17). Various studies, including Devi et al. (18), Harris et al. (19), Karmarkar et al. (20), Sader et al. (21), Pancesso et al. (22), Zhanel et al. (23), Fernandes and Dhanashree (24), and Mihajlović-Ukropina et al. (25), have reported the prevalence of vancomycin resistance in Enterococcal strains to be 10%, 20%, 8.6%, 23.3%, 3.1%, 6%, 23%, 31%, and 31% respectively. According to Figure 1, this bacteria's highest range of contamination was observed in the ICU, maternity and internal ward, which was similar in some research. In Karimzadeh et al. in Shiraz (26), the most contamination by Enterococcus was in the surgical and internal wards, but in Alebouyeh et al. in Tehran, the ICU ward had the highest range of contamination (27). Enterococcal strains are a significant factor in urinary tracheal infection, and they are the third factor of UTI in hospitalized patients, particularly in maternity wards. According to De Francesco et al., in Italy, Enterococci caused 9% of positive cases in outpatients and 12% in inpatients (26).
5.1. Conclusions
Enterococci have become the essential pathogenic bacteria that cause nosocomial infections with multiple-antimicrobial resistance. As mentioned in the results, resistance to ciprofloxacin, ampicillin and vancomycin was widespread among isolates. Therefore, an accurate antibiogram is needed to choose the most effective antibiotics. On the other hand, developing new antibiotics or alternative compounds is necessary.