1. Background
Brucellosis is a zoonotic disease caused by various species of Brucella (1). Brucellosis imposes a severe public health burden on many countries worldwide. People such as farmers, veterinarians, butchers, and laboratory experts are at risk of infection (2). Brucellas are small, gram-negative, intracellular, nonsporulating, nontoxigenic facultative coccobacilli of farm animals such as cows, sheep, pigs, goats, and dogs. According to phenotypic differences and host preferences, ten Brucella species are recognized. Brucella can grow in an aerobic environment; however, some special species, such as Brucella melitensis, B. canis, B. suis, and B. abortus, show better growth in a 10% CO2 environment (3, 4). Brucella melitensis is more common in sheep and goats; B. abortus is more pathogenic in cattle; Brucella swiss's host is pigs; and B. canis is more common in dogs. The prevalence of brucellosis in humans depends on various factors, including dietary habits, husbandry practices, milk processing methods, and environmental sanitation (5, 6). Different serological methods are used to diagnose brucellosis, and most tests depend on detecting anti-Brucella antibodies or bacterial nucleic acid. In Iran, two serological assays are done to confirm the active infection. The Rose Bengal plate test (RBST) is an initial and immediate test performed in laboratories for initial screening. If the sample is positive using this test, it is confirmed by other supplementary tests, such as Wright's and 2ME tests (7). The sensitivity of the RBST is more than 99%, and false-negative results are uncommon. Enzyme-linked immune-sorbent assay (ELISA) is less used than the agglutination tests. The ELISA measures IgG, IgA, and IgM antibodies and has some advantages, such as high sensitivity and better interpretation of the clinical situation. Diagnosis of brucellosis through polymerase chain reaction (PCR) is an ultimately confirmed method, and it can overcome the difficulties of previous serology methods. The main advantages of this test include the highest efficiency, sensitivity, specificity, and diagnostic accuracy. The exact distribution of Brucella in humans is still unknown in many parts of the world, and realizing the species involved in this disease is very important (3, 8, 9).
2. Objectives
Therefore, we investigated Brucella spp. In the sera of people infected with this bacterium in Golestan and Gilan provinces, the main species of this disease was identified, and finally, the possibility of accurate planning for the vaccination of animals at risk was provided.
3. Methods
This cross-sectional study was conducted carried out from 6/2019 to 6/2021 in Golestan and Gilan provinces on patients with clinical symptoms. Blood samples were taken from a total of 2910 patients, referring them to several laboratories for the detection of a possible Brucella infection. Informed consent to participate in the study was obtained from all participants. The collected samples were centrifuged at 2000 × g for 5 minutes, and the serum was stored at -20°C until the test.
3.1. Rose Bengal Test
This test was performed on suspected patients’ sera as an initial screening method. A volume of 30 µL of patient serum was mixed with 30 µL of Rose Bengal antigen (Pasteur Institute of Iran) on a glass slide. The mixture was stirred using a wooden applicator and observed for agglutination within 4 minutes at room temperature. Any visible agglutination was considered positive, and these positive samples were used for confirmatory testing with the Wright test.
3.2. Wright Test
For the Wright agglutination test, patient sera were serially diluted with 0.9% normal saline in test tubes at ratios of 1:20, 1:40, 1:80, and 1:160. Then, 0.5 mL of Wright-specific antigen (Pasteur Institute of Iran) was added to each tube. The tubes were incubated at 37°C for 24 hours. After incubation, the tubes were gently shaken, and the presence of agglutination was observed. A titer of 1:160 or greater was considered a positive result for brucellosis.
3.3. Polymerase Chain Reaction for Identification of Brucella
DNA extraction was done using a DNA extraction kit (GeneAll®, Korea) following the instructions provided in the kit. Initially, 200 μL of binding buffer was added to 200 μL of suspected serum, followed by the addition of 20 μL of proteinase K. The mixture was then incubated for 10 minutes at 70°C. Next, 100 μL of absolute ethanol was added. Next, centrifugation for 1 minute at 8000 × g was done. Subsequently, 600 μL of washing buffer (BW) was added to the samples, followed by centrifugation for 1 minute at 8000 × g. Following that, 700 μL of the second washing buffer (TW) was added to the samples and centrifuged at 8000 rpm for one minute. Another centrifugation step was done again at 13,000 rpm to remove residual buffers for one minute. Finally, 200 μL of elution buffer (AE) was added and allowed to stand for one minute at RT, after which the samples were centrifuged at 13,000 rpm. The extracted DNA was stored at -20°C.
To identify Brucella spp., three sets of primers recommended by the World Animal Health Organization were used (Table 1).
| Primer Set; Gene | Primer Sequences | Amplicon Size (bp) | Source of Genetic Differences | Reference |
|---|---|---|---|---|
| 1 | 450 | IS711 insertion in BMEIO535-BMEI0536 in Brucella strains isolated from marine mammals | (10) | |
| BMI0535 | F: GCGCATTCTTCGGTTATGAA | |||
| BMI0536 | R: CGCAGGCGAAAACAGCTATAA | |||
| 2 | 1071 | Deletion of 25,061 bp in BMEII826-BEII0850 in Brucella abortus | (11) | |
| BMEII0843 | F: TTTACACAGGCAATCCAGCA | |||
| BMEII0844 | R: GCGTCCAGTTGTTGTTGATG | |||
| 3 | 218 | Paint mutation in BMEI0752 in B. melitensis Rev.1 | (12) | |
| BMEI0752 | F: CAGGCAACACCCTCAGAAGC | |||
| BMEI0752 | R: GATGTGGTAACGCACACCAA |
Sequences of Oligonucleotide Primers Used for the Distinction of Brucella Species
The PCR reactions were done in a final volume of 25 µL. Each reaction contained 11 µL of deionized sterile water, 10 µL of Taq DNA Polymerase 2× Mix Red-MgCl2 2 mM (GeneAll, Cat. no. A180301), one picomole of each primer, and two µL of DNA template. The PCR was done by holding at 94°C for 3 minutes and subsequently cycling 38 times at 94°C for 1 min, 55°C for 45 sec, and 72°C for 1 min. Positive and negative controls were included in each PCR run. A known Brucella DNA sample was used as the positive control, and nuclease-free water was used as the negative control to ensure the accuracy and specificity of the amplification process.
The quality and concentration of extracted DNA were assessed using a NanoDrop spectrophotometer (Thermo Scientific™) to ensure an A260/A280 ratio between 1.8 and 2.0, indicating acceptable purity. Gel electrophoresis on 1.1% agarose was also used to confirm DNA integrity.
Primers used in this study were adopted from OIE (World Organisation for Animal Health) guidelines and previous validated studies (Table 1).
The target regions — Deletion of 25,061 bp in BMEII826-BEII0850 for B. abortus and the point mutation in BMEI0752 for B. melitensis Rev.1 — were selected based on their high specificity and discriminatory ability between Brucella spp., as demonstrated in earlier molecular epidemiological reports (13).
All PCR products were electrophoresed using a 1.1% agarose gel, as recommended by the WHO guidelines for Brucella spp. Identification. The molecular weight was determined using a 100 bp DNA ladder (Fermentas, SM0241), and band sizes were interpreted accordingly.
3.4. Statistical Analysis
Microsoft Excel analyzed clinical information about the patients (Ver. 2010). Prism software (GraphPad Software, Inc., CA, USA) was applied for statistical analysis.
4. Results
4.1. Polymerase Chain Reaction
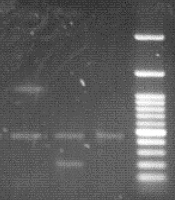
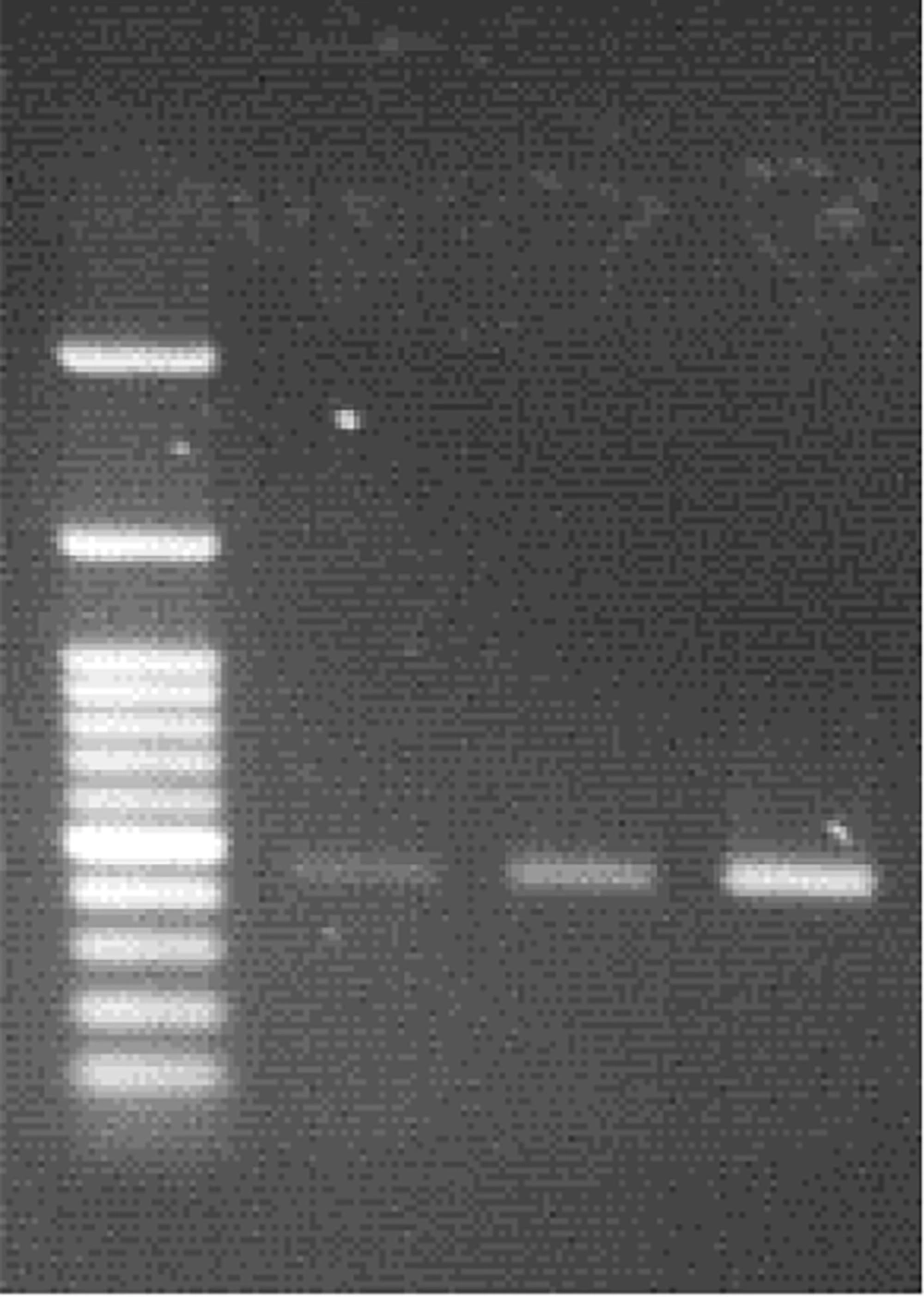
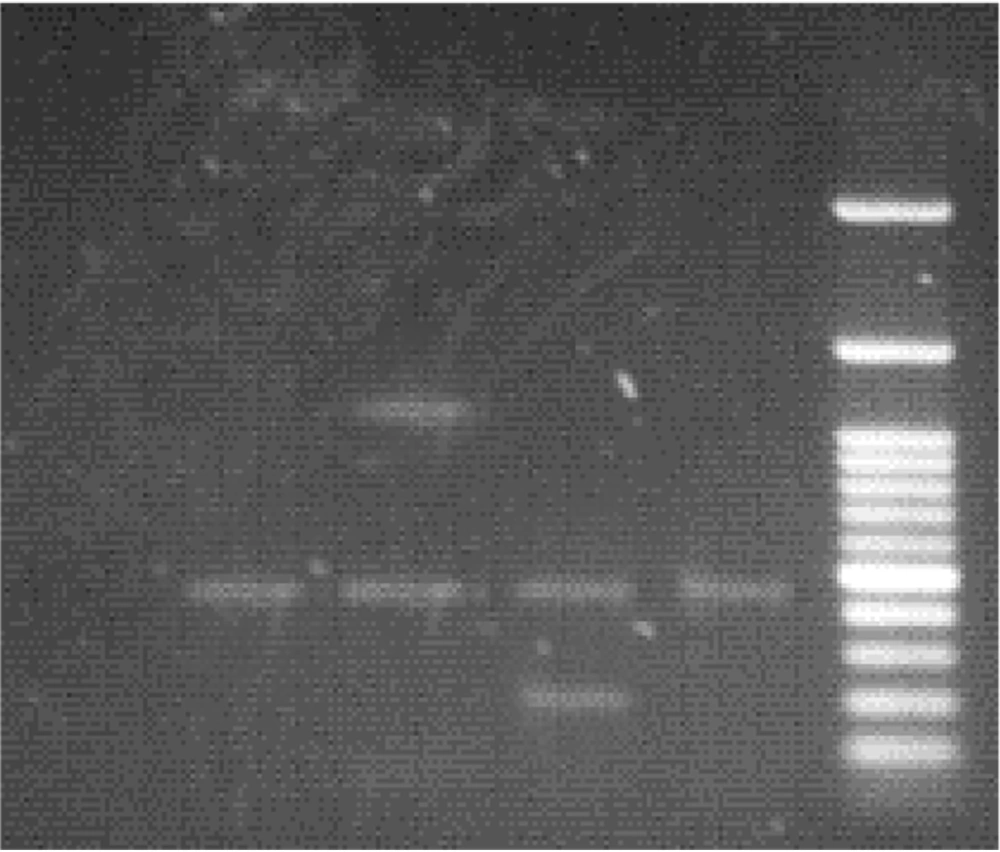
After electrophoresis of PCR products for the detection of Brucella spp, the amplicon size for the Brucella genus was 450 bp, while it was 218 bp for B. abortus and 1071 bp for B. millitensis (Figures 1 and 2).
In Gilan province, 52 (3.56%) and 45 (3.34%) of the evaluated samples were positive on the Rose Bengal and Wright tests, respectively; however, no positive samples were detected after using the PCR test. Of the 52 positive sera in the Rose Bengal test, 3.5% (14) were females, while 3.54% were males.
Of the 1563 patients in the Golestan province, 55 (3.51%) had a positive Rose Bengal test result (including 23 females and 32 males), and 53 (3.39%) were positive on the Wright test. After performing the PCR test, we found that 13 (1.96%) females and 18 (1.99%) males were positive. Out of 31 PCR-positive test samples from Golestan province, 25 were contaminated with B. abortus, and the remaining six were infected with B. melitensis. (Tables 2 and 3).
| Province; Gender | Total Number | Rose Bengal | Wright Test | PCR |
|---|---|---|---|---|
| Gilan | ||||
| Female | 570 | 20 (3.5) | 19 (3.33) | - |
| Male | 777 | 32 (3.54) | 26 (3.34) | - |
| Total | 1347 | 52 (3.86) | 45 (3.34) | - |
| Golestan | ||||
| Female | 661 | 23 (3.47) | 22 (3.32) | 13 (1.96) |
| Male | 902 | 32 (3.54) | 31 (3.43) | 18 (1.99) |
| Total | 1563 | 55 (3.51) | 53 (3.39) | 31 (1.98) |
Demographic and Serological Distribution of Brucellosis Among the Study Population by Province and Gender a
The lack of PCR-positive cases in Gilan province is unclear. Possibilities include lower prevalence, DNA degradation, or differences in sample handling, which warrant further investigation in future research.
The statistical analysis relied solely on descriptive tools without confidence intervals or multivariate models. This limits the robustness of associations drawn from the dataset.
While B. abortus was the most detected species among PCR-positive cases in Golestan, the conclusion should be considered preliminary given the absence of culture confirmation and limited geographic sampling.
The discrepancy between PCR and serological results may be attributed to several factors: Timing of sample collection relative to bacteremia, degradation of bacterial DNA, or false positives in serology due to cross-reactivity.
Potential confounding factors such as occupational exposure, age, and animal contact were not recorded. Their absence limits the interpretation of PCR positivity and should be addressed in future studies.
Although positive and negative controls were included, detailed internal validation of the PCR assay — such as sensitivity, specificity, and detection limits — was not established in this study. This is a recognized methodological gap.
The sample size used for PCR (98 out of 2910 total samples) was limited to seropositive individuals due to cost and logistical constraints. This selective testing may introduce bias and limit the representativeness of PCR findings.
Furthermore, genotypic or strain-level analyses such as MLST or SNP typing were not performed. Future studies incorporating these molecular epidemiological tools would provide greater insight into transmission patterns and strain diversity.
The study did not use culture methods for Brucella detection, which is considered the gold standard. This limitation is acknowledged due to biosafety concerns and limited access to BSL-3 facilities. The reliance on PCR may affect the confirmatory strength of the findings.
5. Discussion
Laboratory tests based on molecular detection are very sensitive and efficient for detecting Brucella spp. Since the principle of these methods is based on the multiplication of specific DNA fragments of the bacteria, they can detect the Brucella in the initial phase of infection. Another advantage of these tests, which has made them preferable to other Brucella detection tests, is their high sensitivity, specificity, and safety of laboratory staff when working with the bacterial genome. In addition, in many cases, when working on old samples in the laboratory, there is a drastic decrease in the serum antibody titer, which interferes with many Brucella detection serologic tests. In contrast, with the PCR test, we can largely overcome this problem and detect the presence of bacteria in these old samples with great accuracy (8, 9).
Iran is endemic to brucellosis, and the average incidence of this disease has been reported at 21 per 100,000. However, the incidence was reported in different regions from 1.5 to 107.5 per 100,000 population (15, 16). The present study aimed to identify Brucella spp. in blood samples of people suspected of brucellosis, referring to diagnostic laboratories in Golestan and Gilan provinces. According to the PCR tests, it was determined that out of 53 positive serum samples in Golestan province, 31 were positive in PCR. The results showed 25 samples infected with B. abortus and six with B. melitensis. This research showed that the PCR test can be used to detect brucellosis and identify the species of Brucella. The percentage of positive cases of brucellosis was 3.39% in the serological test (Wright test) and 1.98% in the PCR test (13, 17).
Unlike reports from other regions of Iran and other countries in the Middle East, in the present study, the predominant species in patients was B. abortus, which is rarely reported in humans from different regions of Iran. In justification of this, it should be said that lifestyle and contact with the type of livestock are the most important factors determining the dominant species of Brucella in a region. One of the important risk factors that plays a role in the contamination of any region with a specific type of brucellosis is the culture of keeping the different kinds of livestock in that region and the dietary habits of the people of that country (18, 19).
In many regions of Iran, due to the nomadic lifestyle of the people, the most livestock kept are sheep and goats. Therefore, of course, the most contact and use of livestock products in these areas is from sheep and goats. Sheep and goats are usually known as the main hosts of B. melitensis, and it is obvious that most infections with Brucella spp. are related to B. melitensis (20, 21).
Most of the animals kept in Golestan province in a traditional or semi-industrial way are cattle and sheep. Unfortunately, because of the neighboring countries, the illegal entry of smuggled animals from other countries has caused a high level of B. abortus prevalence among the humans of these regions. In addition, one of the other factors that increase the majority of B. abortus in this province is the high consumption of non-pasteurized animal products, which is due to various factors, such as the lack of suitable industrial factories in this province and the general culture of the people of this region, which results in less consumption of pasteurized and processed factory products.
Ali Ahmadi et al. found that 11.3% of samples tested positive using PCR. The results of the Wright and 2-ME tests in their study were consistent with the PCR results. They concluded that sheep and goat brucellosis is more prevalent in Sistan and Baluchestan provinces than in other parts of Iran (14). The present study found a similar correlation in Golestan province, but not in Gilan province.
Dadar et al. conducted phenotypic and molecular analysis on various Brucella isolates, revealing that both B. melitensis and B. abortus play a role in the prevalence of human brucellosis in Iran. The B. melitensis isolates mainly consisted of MLST-9 ST8 and ST7 genotypes, with some instances of ST102. The B. abortus isolates were classified into two widely distributed MLST-9 genotypes (ST1 and ST2) (22).
A meta-epidemiological study published by Dadar et al. revealed that the most studied species in Iran were cattle, followed by sheep, goats, camels, and buffalo. The most commonly prevalent Brucella spp. found in livestock were B. melitensis, B. abortus, mixed infections of B. melitensis and B. abortus, and the vaccine strain of B. melitensis Rev1. The PCR-based tests were the most frequently used method to detect Brucella spp., while indirect ELISA showed the highest prevalence of Brucella-positive cases (69%). Interestingly, the prevalence of brucellosis was significantly higher in females (10.91%) compared to males (8.23%) (23). The results of this study indicate that in Golestan province, the most commonly consumed livestock products are cow and sheep, leading to a higher incidence of human infection with B. abortus in this area. Additionally, a key factor contributing to the spread of this bacterium among the human population is the consumption of unpasteurized animal products. This behavior is influenced by various factors, including the absence of adequate industrial facilities for the production of animal products, a risk factor that is particularly evident in Golestan province. In 2023, Aminzadeh et al. did not recommend isolating bacteria from sheep milk due to its low sensitivity, time-consuming nature, and associated risks. Instead, they suggested using modified RBT as a screening test because of its diagnostic accuracy, higher sensitivity, and accuracy. They also recommended using real-time PCR as the gold standard test for detecting brucellosis in sheep milk (24). In the present study, the frequency of positive cases obtained by PCR test was lower than the positive cases obtained by serological tests, which shows that the false positive cases of serological tests were more than those of PCR.
This study has several limitations. First, although PCR provides high specificity, false negatives may occur due to low bacteremia or DNA degradation. Second, the study was limited to only two provinces, which restricts generalizability. Third, the lack of culture confirmation may impact the interpretation of the results. Finally, resource constraints limited extensive genotyping or real-time PCR validation.
5.1. Conclusions
This study demonstrated that B. abortus, rather than the commonly reported B. melitensis, is the predominant species causing human brucellosis in Golestan province. This finding highlights the need for species-specific surveillance and region-tailored control measures, including targeted vaccination strategies and improved regulation of unpasteurized animal product consumption.