1. Background
The type and distribution of melanin in human skin play crucial roles in determining skin pigmentation (1, 2). These factors have been linked to latitude, UV radiation exposure, lifestyle, diet, metabolism, and genetics. The genetic component plays a significant role in the biosynthesis of 2 types of melanin (pheomelanin and eumelanin), which have been linked to skin cancer (1, 2). One genetic factor that has been linked to the development of melanoma is the solute carrier family 45 member 2 (SLC45A2) gene, located on chromosome 5p13.2. This gene consists of 7 exons and encodes a protein called the membrane-associated transporter protein (MATP) that plays an important role in melanin synthesis in melanosomes (3, 4). The protein encoded by the SLC45A2 gene is a member of the K+/Na+/Ca2+ exchanger family of transmembrane proteins and is thought to regulate calcium concentration, pH, and ionic homeostasis within melanosomes (5).
Melanosomal pH and ionic homeostasis are crucial for melanin synthesis by regulating tyrosinase activity and melanosome development. During the initial stages of melanosome development, an acidic pH helps L-DOPA stabilization by preventing auto-oxidation. As melanosomes maturation progresses, increasing the pH optimizes tyrosinase activity and enhances melanin production. A change in melanosomal pH can alter melanin production (4, 6).
Variants in the SLC45A2 gene have been linked to darker hair, eye pigmentation, and melanoma in populations worldwide, including North America, Asia, Europe, Africa, and specific ethnic groups such as Japanese, South African European, Chinese, French, Italian, and Brazilian (6-9). These variants cause a lack of melanin, resulting in albinism, especially oculocutaneous albinism type 4 (OCA4) (10, 11). In Colombia, similar genetic associations have been identified, with some variants in different genes [such as cyclin dependent kinase inhibitor 2A (CDKN2A) and cyclin dependent kinase 4 (CDK4)] linked to melanoma development; in addition, SLC45A2 has been considered as a moderate and low penetrance germline variant affecting melanoma risk (12-14).
The p.Glu272Lys variant occurs in exon 3 due to a G > A transition, leading to an amino acid change at codon 272. The p.Phe374Leu variant is caused by a G > C transversion in exon 5, resulting in an amino acid change at codon 374. These variants have a significant impact on the functionality of the protein, as they can eliminate or create protein binding sites (15). However, the prediction of pathogenic variants in genes can aid in understanding their roles in various disease mechanisms (16, 17). In silico methods are used to predict whether these variants have a deleterious or neutral effect by analyzing protein structure (18, 19).
2. Objectives
As the aforementioned variants are associated with different diseases and melanoma, the objective of this study was to identify the association between SLC45A2 variants with the susceptibility to melanoma development in the Colombian population.
3. Methods
3.1. Study Population
This analytical observational case-control study was conducted at the Hospital Universitario Centro Dermatologico Federico Lleras Acosta between 2018 and 2020. The case group was comprised of patients enrolled during their visits to the hospital’s tumor clinic, who were then reviewed by an oncology dermatologist. On the other hand, the control group consisted of healthy patients screened by the dermatologist for any melanoma-suggestive lesions. The sample size was calculated based on the frequency of the minor allele using the Kelsey method with a case-control ratio of 1: 2. All clinical, pathological, and sociodemographic characteristics were collected by the medical and research team. The study was approved by the Research Ethics Committee of Hospital Universitario Centro Dermatologico Federico Lleras Acosta. Genomic DNA was extracted from peripheral blood samples using the QIAamp DNA Mini and Blood Mini Handbook kit and diluted to a final concentration of 20 ng/uL.
3.2. Genotyping SLC45A2 Variants
The results showed that the two SLC45A2 variants (p.Glu272Lys rs26722 and p.Phe374Leu rs16891982) were associated with melanoma. 10 samples of melanoma cases and 10 samples of a control group were analyzed using the Sanger method to serve as a control for the subsequent analysis, which was performed using the Big Dye Terminator and BI PRISM 3130xl Genetic Analyzer (Applied Biosystems). The PCR conditions and SLC45A2 primers (p.Glu272Lys 5’-CTGCATTGCCAGCTCTGGAT-3’, 5’-CCTCAGGACCCTCCATTGTC-3’ and p.Phe374Leu 5’-GGAGAGAGAAAGACTTACAAGAATAAA-3’, 5’-TCCACAGAGTTTCTCATCTACGA-3’) were described in a previous study (13). All SNPs were genotyped using real-time (qPCR) and analyzed with high-resolution melting analysis (HRM). The conditions used for this process were also described in previous study (12, 13).
3.3. In Silico Analysis of Novel Mutations
To summarize, various online tools and software were used to obtain information about the SLC45A2 gene, including its sequence and structure. These tools were used to explore the domain architecture of the protein encoded by the gene, predict the impact of variants on the protein’s function and stability, and compare the 3-dimensional structure of different variants. These tools include GeneCards, GenBank, UniProt, SMART, ROBETTA, ChimeraX, Polyphen-2, Pmut, and PROVEAN. The goal of these analyses was to determine the pathogenicity of novel mutations and the impact of amino acid substitutions on the protein’s function (20).
3.4. Statistical Analysis
The statistical analysis for this study was conducted using Stata version 16 (StataCorp LLC, College Station, Texas, USA). All odds ratios (OR) were reported with their 95% CIs. P values less than 0.05 were considered statistically significant. The association between genetic factors and melanoma was tested using the Hardy-Weinberg equilibrium, the chi-square test, and OR with CI; the data were analyzed using logistic regression. The analysis of genotype, haplotypes, and allele frequency was performed using SNPStats software (http://bioinfo.iconcologia.net/SNPstats) from the Catalan Institute of Oncology in Barcelona, Spain. The association was tested using 4 different models (i.e., recessive, dominant, additive, and codominant genotypic models) and was adjusted for both sex and the family history of cancer.
4. Results
The study included 85 cases and 166 controls. The average age was 59 for cases and 57 for controls (54% of cases and 54% of controls were female). Most participants in both groups were born in the central region of Colombia (Cundinamarca) and had fair or dark features. A history of familial melanoma was present in 8% of cases. More cases had more than 100 moles compared to controls. Melanoma subtypes included 36% lentigo maligna melanoma (LMM), 24% acral lentiginous melanoma (ALM), 24% superficial spreading melanoma (SSM), and 15% nodular melanoma (NM). Pathologic studies showed that 41% of cases had Clark level IV, and 25% had a Breslow thickness of more than 1 mm, of whom 51% had mitoses and 21% had ulceration (13).
4.1. Analysis Between SLC45A2 Variants and Phenotypic Characteristics
The analysis of SLC45A2 variants showed a correlation with the phenotypic characteristics of melanoma. The p.Glu272Lys variant was present in 42% of cases and 33% of controls, while the p.Phe374Leu variant was present in 42% of cases and 48% of controls. The cases with the p.Glu272Lys variant had a higher percentage of LMM (17.65%), IV Clark level (17.56%), and Breslow scale > 4.0 mm (9.41%); they were mostly located on the head and neck (21.18%). Similarly, the cases with the p.Phe374Leu variant had a higher percentage of LMM (16.47%), IV Clark level (20%), and Breslow scale > 4.0mm (11.76%); they were mostly located on the head and neck (18.82%; Table 1).
| Feature | P.Glu272Lys | P.Phe374Leu | ||||||
|---|---|---|---|---|---|---|---|---|
| CASES (N = 85) | Controls (N = 166) | CASES (N = 85) | Controls (N = 166) | |||||
| Presence (%) | Absence (%) | Presence (%) | Absence (%) | Presence (%) | Absence (%) | Presence (%) | Absence (%) | |
| Age | ||||||||
| Mean | 59 | 59 | 58 | 58 | 60 | 58 | 58 | 57 |
| Sex | ||||||||
| Women | 21 (25) | 25 (29) | 29 (17) | 60 (36) | 20 (24) | 26 (31) | 44 (27) | 45 (27) |
| Melanoma subtype | ||||||||
| SSM | 8 (9) | 12 (14) | - | - | 8 (9) | 12 (14) | - | - |
| NM | 4 (5) | 9 (11) | - | - | 5 (6) | 8 (9) | - | - |
| ALM | 9 (11) | 12 (14) | - | - | 11 (13) | 10 (12) | - | - |
| LMM | 15 (18) | 16 (19) | - | - | 14 (16) | 17 (20) | - | - |
| Clark Level | ||||||||
| Negative | 17 (20) | 16 (19) | - | - | 17 (20) | 16 (19) | - | - |
| I - II | 3 (3) | 0 (0) | - | - | 2 (2) | 8 (9) | - | - |
| III | 1 (1) | 1 (1) | - | - | 1 (1) | 1 (1) | - | - |
| IV - V | 15 (18) | 25 (30) | - | - | 18 (21) | 22 (26) | - | - |
| Breslow scale (mm) | ||||||||
| Non-reported | 17 (20) | 15 (18) | - | - | 16 (19) | 16 (19) | - | - |
| ≤ 1.0 | 4 (5) | 7 (8) | - | - | 4 (5) | 7 (8) | - | - |
| > 1.0 - 2.0 | 1 (1) | 8 (9) | - | - | 2 (2) | 7 (8) | - | - |
| > 2.0 - 4.0 | 6 (7) | 6 (7) | - | - | 6 (7) | 6 (7) | - | - |
| > 4.0 | 8 (9) | 13 (15) | - | - | 10 (12) | 11 (13) | - | - |
| Location | ||||||||
| Trunk | 5 (6) | 7 (8) | - | - | 7 (8) | 5 (6) | - | - |
| Head and neck | 18 (21) | 19 (22) | - | - | 16 (19) | 21 (25) | - | - |
| Upper-lower extremities | 4 (4) | 11 (11) | - | - | 4 (5) | 11 (13) | - | - |
| Hands and feet | 9 (11) | 12 (14) | - | - | 11 (13) | 10 (12) | - | - |
| Phototype | ||||||||
| 3 | 26 (31) | 33 (39) | 40 (4) | 76 (46) | 31 (36) | 28 (33) | 63 (38) | 53 (32) |
| 4 | 6 (7) | 4 (5) | 8 (5) | 9 (5) | 3 (4) | 7 (7) | 8 (5) | 9 (5) |
| Eye color | ||||||||
| Black or dark brown | 19 (22) | 25 (29) | 42 (25) | 85 (51) | 20 (24) | 24 (28) | 63(38) | 59 (36) |
| Light brown | 12 (14) | 18 (21) | 11 (7) | 16 (10) | 11 (13) | 19 (22) | 13(8) | 14 (8) |
| Green - blue | 5 (6) | 6 (7) | 3 (2) | 13 (8) | 7 (7) | 4 (5) | 5(3) | 11 (7) |
| Hair color | ||||||||
| Black or dark brown | 26 (31) | 29 (34) | 48 (29) | 87 (52) | 30 (35) | 25 (29) | 69 (42) | 66 (40) |
| Light brown - red | 10 (12) | 20 (23) | 8 (5) | 22 (14) | 8 (9) | 22 (26) | 12 (70) | 19 (12) |
| Nevus | ||||||||
| ≤ 50 | 33 (39) | 38 (45) | 54 (33) | 106 (64) | 33 (39) | 38 (45) | 81 (49) | 79 (48) |
| 50 - 100 | 2 (2) | 9 (11) | 2 (1) | 3 (3) | 4 (5) | 7 (8) | 0 | 5 (3) |
Abbreviations: LMM, lentigo maligna melanoma; ALM, acral lentiginous melanoma; SSM, superficial spreading melanoma; NM, nodular melanoma.
4.2. SLC45A2 Variants Analysis
The analysis of allele and genotypic results for the rs26722 and rs16891982 variants in the SLC45A2 gene showed that the C allele of the rs26722 variant acted as a protective factor for melanoma in the population (OR, 0.656; 95% CI, 0.44-0.96; P = 0.029), while the G allele of the rs16891982 variant was associated with the development of melanoma (OR, 2.447; 95% CI, 1.488 - 4.025; P = 0.0003; Table 2). No statistical differences were observed between the case and control groups in various genetic models (including dominant, recessive, codominant, overdominant, and log-additive models) after adjusting for sex and the family history of cancer (Appendix 1).
| Genotype/Allele | Cases (n = 85 %) | Control (n = 166 %) | OR | 95% CI | P Value |
|---|---|---|---|---|---|
| SLC45A2 (rs26722 p.E272K) | |||||
| T/T | 36 (42) | 50 (30) | 1 | Reference | |
| T/C | 0 (0.0) | 6 (4) | 0.144 | (0.008 - 2.594) | 0.128 |
| C/C | 49 (58) | 109 (66) | 0.711 | (0.416 - 1.217) | 0.213 |
| C | 98 (58) | 224 (67) | 0.656 | (0.44 - 0.96) | 0.029 a |
| SLC45A2 (rs16891982 p.L374F) | |||||
| C/C | 47 (55) | 84 (51) | 1 | Reference | |
| G/C | 37 (44) | 79 (48) | 0.848 | (0.501 - 1.43) | 0.541 |
| G/G | 1 (1) | 2 (1) | 0.976 | (0.08 - 10.92) | 0.984 |
| G | 39 (23) | 83 (25) | 2.447 | (1.488 - 4.025) | 0.0003 a |
Abbreviations: SLC45A2, solute carrier family 45 member 2; OR, odds ratio.
a Significant P value.
4.3. Haplotype Analysis
The analysis of the haplotypes adjusted for sex and phototype showed that the CG haplotype was a risk factor for melanoma (OR, 2.71; 95% CI, 1.20 - 6.13; P = 0.018; Table 3).
| P.E272K | P.L374F | Frequency | OR (95% CI) | P Value |
|---|---|---|---|---|
| C | C | 0.5372 | 1 | - |
| T | C | 0.2182 | 1.09 (0.67 - 1.76) | 0.73 |
| T | G | 0.1407 | 0.74 (0.39 - 1.40) | 0.35 |
| C | G | 0.104 | 2.71 (1.20 - 6.13) | 0.018 a |
Abbreviation: OR, odds ratio.
a Significant P value.
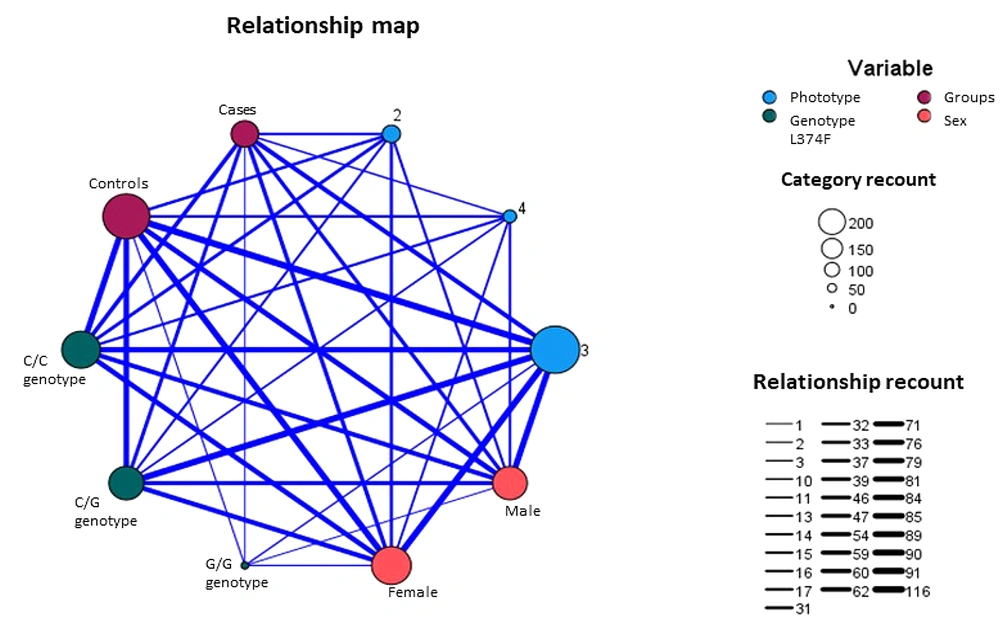
To analyze the relationship between clinical factors and genetic factors in melanoma, clinical factors were divided into 2 groups (phototype I-II vs phototype III-IV, light eye color vs black eye color, red/blond/brown hair color vs dark-brown / black hair color, and nevus count < 50 vs nevus count > 50) and analyzed in relation to the genetic factors (rs26722 and rs16891982). It was found that the p.Phe374Leu variant acted as a protective factor in the population with phototype III-IV and dark-brown and black hair (OR, 0.380; P = 0.004; and OR, 0.459; P = 0.011, respectively; Appendix 2). We conducted a mapping of the relationships among the variables of phototype, genotype (in both the cases and control groups), and the sex of the analyzed population. Our finding revealed a strong correlation between these variables, particularly the groups defined by phototype, the present of the p.Phe374Leu variant, and the sex of the patients (Figure 1).
The relationship diagram between phototype, sex, groups, and p.Phe374Leu. Category recount circle’s sizes represent the rounded value for members of each category. The relationship recount line’s thicknesses represent the rounded value for the member of a category that is related to another one. Blue circles represent the phototype variable. Red circles represent groups (cases and controls). Peach circles represent sex (male and female). Green circles represent the genotype for the rs16891982 variant.
4.4. Modeling of the MATP Protein Structure and Validation
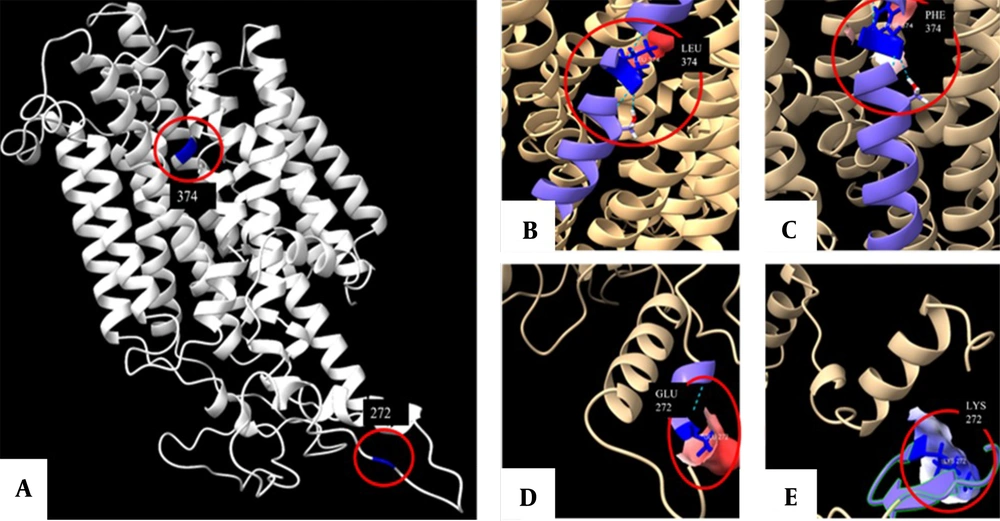
Information about the gene and protein sequence of SLC45A2 was obtained from the GenBank and UniProt databases. The intramembrane and cytoplasmic domains were identified using SMART software (Appendix 3). The protein structure was predicted using Robetta software and visualized using ChimeraX software (Figure 2). The variants were found to have an impact on the protein’s biological function; the p.Phe374Leu variant had a pathogenic effect, while the p.Glu272Lys variant did not have any pathogenic effect (Appendix 4).
The structure of the membrane-associated transporter protein (MATP) prediction in ChimeraX software. This structure was predicted using the Protein Data Bank (PDB) formats obtained by the ROBETTA tool. Panel A show a front view of the protein with positions 272 and 374 highlighted in blue. Panels B and C display the front view of the L374 and F374 variants, respectively. Furthermore, panels D and E provide a detailed view of each amino acid, including the hydrogen bonds it forms (represented by dotted lines), as well as a simulation of the surface based on the electrostatic Coulomb potential. The potencial varies from red (indicating a residue with a higher negative potential) to blue (indicating a more positive potential).
5. Discussion
Skin pigmentation is closely linked to susceptibility to melanoma. The presence of pheomelanin is a risk factor, and the presence of eumelanin is a protective factor (21). By studying genetic variants in different populations, we can understand the adaptive genetic characteristics of a population in relation to a specific geographic region and establish a correlation between an individual’s genotype and phenotype to identify predictors of susceptibility to disease.
The variants of the SLC45A2 gene have been previously linked to susceptibility to melanoma due to their varying population frequencies and association with dark hair, dark skin, and dark eye pigmentation (22, 23). In this study, p.Glu272Lys and p.Phe374Leu variants were investigated. The results showed that the presence of the C allele of the p.Glu272Lys variant acted as a protective factor, while the presence of the G allele of the p.Phe374Leu variant was found to be a risk factor for melanoma. Previous studies have shown that the p.Phe374Leu variant is a risk factor for melanoma in individuals of Spanish origin, as well as a protective factor when adjusted for other genetic and phenotypic factors in French, Italian, and Spanish populations (2, 3).
The p.Phe374 allele is fixed in European populations, while the p.Leu374 allele is fixed in African populations; however, it is not associated with skin pigmentation (24). The presence of phenylalanine at position 374 of the protein encoded by SLC45A2 makes it less stable than the p.Leu374 variant and leads to reduced levels of the protein, resulting in less production of eumelanin and lighter pigmentation (25). Therefore, the presence of p.Phe374 is a risk factor for melanoma in populations with light phototypes. However, a negative selective pressure has been shown in individuals in Northern Europe for the ancestral G allele (24, 26).
The present study analyzed the frequency of 2 variants (p.Glu272Lys and p.Phe374Leu) in the SLC45A2 gene in a population living in equatorial regions with a mix of African, European, and indigenous ancestry. The frequency of the G allele (p.Phe374Leu) in this population was found to be similar to that found in a population from northern Brazil but much higher than that found in European regions and lower than that found in the African population (16, 24, 27). The T allele at SNP c.814C > T (p.Glu272Lys) was found to have a frequency similar to that found in the Japanese population; however, in European populations, it is considered a protective factor (16, 24). The results of this study also showed that even though genotype analysis did not show an association with melanoma, the presence of the CG haplotype is a risk factor when adjusted for phototype and age. The results suggest that the protective effect of these variants may vary across different populations and that further research is needed to fully understand the relationship between these variants and melanoma risk (28, 29).
In conclusion, the analysis of SLC45A2 variants and their correlation with phenotypic characteristics in a population of melanoma patients and controls showed that the p.Glu272Lys variant was associated with a subtype of cutaneous melanoma, specifically LMM, while the p.Phe374Leu variant was not found to be associated with any specific subtype. The C allele of the rs26722 variant acted as a protective factor for melanoma in the analyzed population, and the G allele of the rs16891982 variant was associated with the development of melanoma. The CG haplotype was also found to be a risk factor for melanoma.
The strengths of the study include providing information about the genes involved in or associated with melanoma development. These genes are considered high, moderate, or low-penetrance genes based on the population characteristics. The Colombian population is a mix of Europeans, Native Americans, and Africans. Understanding our genetic profile allows us to understand why the behavior of melanoma in Colombia is different from other populations around the world.
The limitations of the study include the small sample size. Further studies with larger sample sizes are needed to increase the statistical power between the studied variants and their association with melanoma development.