1. Context
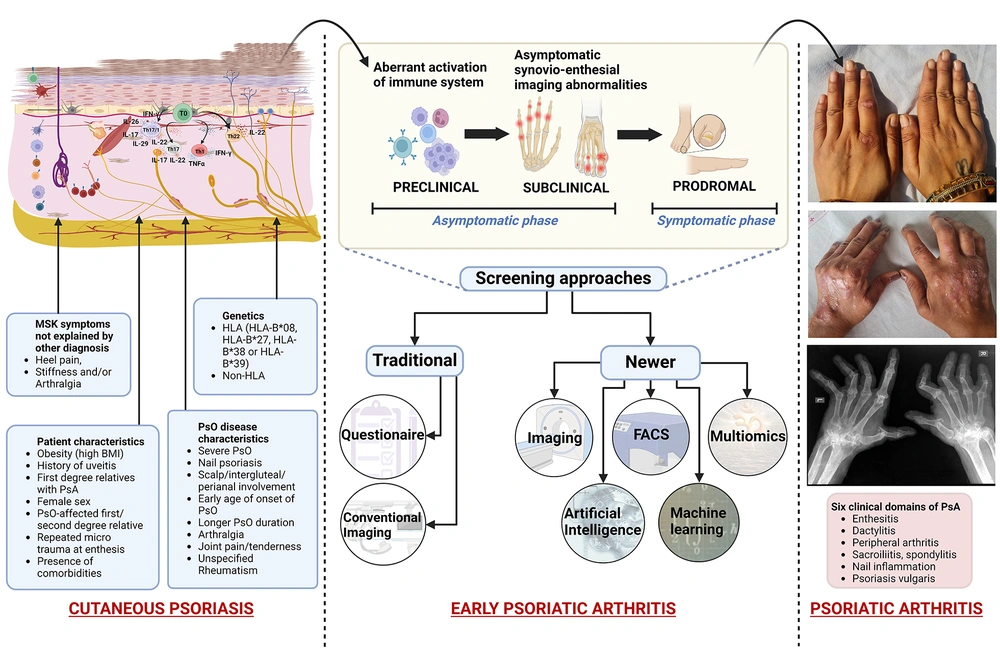
Psoriatic arthritis (PsA) is a chronic immune-mediated disease with a variable prevalence of 0.05% to 0.25% in the general population and 6% to 42% in psoriasis (PsO) patients (1-7). This seronegative, irreversible inflammatory arthritis displays a broad clinical spectrum, ranging from mild oligoarthritis to mutilating polyarticular forms, along with enthesitis and dactylitis (4, 8). Such heterogeneous clinical presentation frequently impedes the early detection of PsA. The shift in the site of disease activity from the skin to the joints (early or developing PsA) is punctuated by mildly symptomatic to almost asymptomatic (clinically quiet) phases, namely the preclinical phase (aberrant immune activation from the skin), subclinical phase (imaging findings without appreciable clinical symptoms, i.e., asymptomatic synovio-enthesial imaging abnormalities), and prodromal phase (arthralgia and fatigue without synovitis and/or enthesitis on examination) (8-10).
The time interval in the progression of PsA from PsO creates an “early” transition window, which can be exploited for identification, interception, and intervention to halt joint disease progression (2, 4, 6, 8-10). Early patient identification will enable dermatologists to timely refer at-risk patients to rheumatologists, who are otherwise consulted when significant, irreversible joint damage has already occurred (11). Currently, there are no specific diagnostic markers for early PsA (10, 12, 13). Prevention/reduction of PsA development has now been suggested as a new clinical endpoint in PsO trials. Since extensive inflammatory and musculoskeletal damage makes early therapeutic intervention quite challenging, there is an urgent need to identify distinct and reliable biomarkers for the early and timely prediction of possible transition to PsA in PsO (14).
2. Evidence Acquisition
A comprehensive literature search was conducted on the PubMed, Web of Science, EMBASE, and SCOPUS databases using keywords such as 'investigation of psoriatic arthritis,' 'early psoriatic arthritis,' 'subclinical PsA,' ‘multiomics in PsA,’ and ‘artificial intelligence in PsA.' All relevant articles were reviewed to investigate current modalities for early PsA, with a special emphasis on newer serological biomarkers and recent research on the role of machine learning and artificial intelligence in the development of PsA.
3. Results
The current predictive/diagnostic protocol in PsA involves extensive clinical questionnaires based on genetic and environmental components in medical history, physical examination, along with biochemical (immunological) and imaging (radiological) studies (15).
3.1. Clinical (Long-Term) Predictors
These include early age of onset of PsO, severe cutaneous disease, inverse PsO, nail pitting, obesity, PsO-affected first/second-degree relative, and uveitis as predictive clinical markers (5, 6, 9, 10, 16, 17). Non-specific musculoskeletal symptoms, such as joint pain/tenderness and arthralgia, might be suggestive, yet not completely diagnostic of PsA (6). Enthesitis (inflammation at the site of attachment of tendons, ligaments, and joint capsules to bone) is a prominent pathological sign, while peripheral oligoarthritis is often the most frequent pattern of PsA presentation (4). Dermatologists also utilize PsA screening and evaluation (PASE), psoriasis epidemiology screening tool (PEST), Toronto psoriatic arthritis screen (ToPAS), updated ToPAS II, early arthritis for psoriatic patients (EARP), CONTEST questionnaire, simple psoriatic arthritis screening (SiPAS), and modified Rheumatic Disease Comorbidity Index as screening tools (1, 6). Latest studies have explored graphical models, such as Bayesian Network correlating clusters of patient symptoms (18). However, robust utilization of these clinical tools is limited by their overall low sensitivity for early PsA (8). In practice, an objective evaluation of patient symptomatology is often difficult due to the non-specific nature of the musculoskeletal symptoms, limited data for their accurate definition, and diverse views regarding the relative significance that should be assigned to the proposed risk factors. This leads to an overall limited ability to precisely predict true disease progression (8).
3.2. Genetic (Long-Term) Predictors
Both PsO and PsA show characteristic genetic markers in HLA (human leukocyte antigen) regions (genes for antigen presentation and immune recognition) and non-HLA regions (genes for inflammation and immune activation, including cytokine expression, intracellular signaling, and T-cell effector function). Many of these genetic associations are not identical, and certain genes are associated with specific phenotypes in PsA (2, 17, 19). For instance, PsA has a weaker association with HLA-B*57:01 and HLA-C*06:02 (psoriasis susceptibility region 1 or PSORS1) and lacks association with interleukin (IL)-12B (5). Similarly, HLA-B*07, HLA-B*08, HLA-B*27, HLA-B*38, HLA-B*39, and HLA-C*12 are strong genetic risk factors for PsA development (8, 14, 19). Further, HLA-C*06 has been shown to delay PsA onset, thus conferring some protection to at-risk patients. Major histocompatibility complex class I chain-related A (MICA) alleles are also implicated in PsA (10, 15, 16). Non-HLA genes include interleukin 23 receptor (IL-23R), tumor necrosis factor alpha-induced protein 3 (TNFAIP3), human c-rel gene (REL), FBX19, and protein tyrosine phosphatase non-receptor type 22 (PTPN22) (10, 16). Other genes exclusively associated with PsA include B3GNT2, IL, KIR2D, KIF3A, and PTPN22 (14). Methylated DNA and CpG markers are additional predictive markers (20).
3.3. Imaging/Radiological (Short-Term) Predictors
PsA-affected joints can show diverse structural (new bone formation, gross joint destruction, erosions, "pencil-in-cup" deformity, joint space narrowing) and inflammatory (synovitis, tenosynovitis, periarticular inflammation, bone marrow edema) changes, driven by altered ‘osteoremodelling’ with increased pathological bone resorption and aberrant periarticular new bone formation (6, 8, 11). While structural features are mostly detectable by conventional radiography, inflammatory features are better identified via magnetic resonance imaging (MRI) (6, 9, 11). Musculoskeletal ultrasound (MSUS), MRI, and Positron emission tomography (PET) further identify subtle preclinical vascularization changes and subdermal inflammation as the earliest changes in PsA transition (5). Positron emission tomography reliably differentiates synovitis from PsA enthesitis. Fluorescence optical imaging (FOI) has recently emerged as successful at accurately detecting hypervascularization during PsA transition that may evade detection by MRI (10, 16). More advanced imaging modalities include High-resolution peripheral quantitative CT (HR-pQCT) (for structural entheseal lesions demonstrating low cortical volumetric bone mineral density), Shear-Wave Elastography using Gray Scale (GS) ultrasound and Power Doppler (for Achilles tendinopathy), and bone scintigraphy (for subclinical joint involvement in PsA without clinical arthropathy) (6, 8).
3.4. Biochemical/Serological (Short-Term) Predictors
Due to altered inflammatory cascade and osteoremodelling, certain related biomarkers (Table 1) have been investigated for their potential role in predicting PsA transition (18-24).
| Variables | Predictors |
|---|---|
| 1. Markers of inflammation | CRP, high-sensitivity-CRP |
| IL-6 | |
| Serum calprotectin | |
| S100A8/S100A9 | |
| 2. Bone and cartilage turnover biomarkers | |
| Catabolic markers (quantifying collagen/bone ECM breakdown) | PRO-C1 (propeptide) |
| PRO-C3 | |
| PRO-C4 | |
| COMP | |
| Anabolic markers (estimating formation peptides of bony collagens | C1M |
| C2M | |
| C3M | |
| C4M | |
| C10C | |
| Others | Dickkopf-1 |
| Sclerostin | |
| RANKL | |
| Bone alkaline phosphatase | |
| OPG | |
| 3. Proteases | MMP-3 |
| Stromelysin-1 | |
| 4. Proteins | |
| Autoantibodies | Anti- MCV |
| Anti-PsA peptide | |
| Anti-N-RAP | |
| Anti-ADAMSTS5 | |
| Anti-cathelicidin LL37 (LL37) | |
| Carbamethylated anti-LL37 | |
| IgA anti-oxidized collagen type II (oxPTMCII) | |
| Others | C-X-C motif chemokine ligand10 (CXCL10) |
| Soluble IL-2 | |
| ITGB5 | |
| M2BP | |
| Myeloperoxidase |
Biochemical/Serological (Short Term) Predictors
The predictor models currently in application chiefly encompass inputs from patient clinical history, radiological details, and laboratory serological investigations for a definitive answer. While patient history and evaluation of clinical features may not always reveal the absolute features of transition to PsA, the inflammatory and musculoskeletal damage depicted in imaging studies is usually quite extensive by the time patients are referred for rheumatological intervention, thus derailing early therapeutic intervention to halt the PsO-to-PsA transition (14). Among the serological markers, some studies have demonstrated their strong, positive, but not predictive role. However, other studies have shown conflicting results, thus limiting their utility by clinicians (2, 10). So far, only CXCL-10 has emerged as the marker with definite positive predictive value (2). Negative traditional arthritic markers such as rheumatoid factor (RF) and/or anti–cyclic citrullinated peptide antibodies (ACPA) limit their diagnostic role in PsA (10). Due to the paucity of a single specific disease predictor/indicator, there is a recent thrust on devising a multi-specialty approach incorporating soluble biochemical markers, imaging techniques, and multi-omics in PsO-to-PsA transition (Figure 1). Furthermore, digital investigative medicine and artificial intelligence are the latest research hotspots, as explained below.
PsO-to-PsA continuum depicting predictive indicators (clinical and imaging features with risk factors) for progression to PsA. “Created with BioRender.com”. PsA, psoriatic arthritis; PsO, psoriatic arthritis; 'HLA, human leukocyte antigen; MSK, musculoskeletal; BMI, body mass index; FACS, fluorescence-activated cell sorting analysis.
3.5. The “Omics” Technology
This technology allows for deciphering complete gene profiles (genomics and epigenomics), mRNA profiles (transcriptomics), protein profiles (proteomics), metabolite profiles (metabolomics), and lipid profiles (lipidomics) in a specific biological sample (14, 25-28). The EU-funded HIPPOCRATES project is one such collaboration aimed at analyzing and combining multiple molecular markers with conventional screening tools (13).
3.5.1. Genomics
Both PsO and PsA possess shared as well as distinct genetic pools (5, 8, 14, 18). Genome-wide genotyping arrays, Genome-wide association studies (GWAS), and immunoprecipitation techniques have shortlisted > 20 additional non-HLA loci, some of which are exclusive to PsA (14). Additionally, epigenetic events such as DNA methylation, histone modification, and small regulatory RNAs are being analyzed. The Oxford Biodynamics EpiSwitch platform has focused on specific 5-marker chromosome conformation signatures (sequences controlling 3D genomic structure) in genomic regions of IL-21R, IFNAR1, IL-23, IL-17A, and CXCL13 (14, 19, 25). Faster, cheaper, and easy-to-automate Next-generation sequencing platforms are also being explored (13).
3.5.2. Proteomics
Analysis of proteins in peripheral blood, serum, plasma, synovial fluid, urine, or skin specimens is being attempted, targeting > 3000 proteins using different selectivity concepts and amplification capabilities (25-27). Synovial fluid mass spectrometry has revealed 12 candidate PsA markers, including MMP3, S100A9, and CRP. Periostin (related to cell adhesion molecules and angiogenesis) has also been identified (14, 19, 25). More recently, spatial immunoproteomics approaches defining specific antigens in the epidermis and dermis are being studied to better understand humoral and cell-mediated immune mechanisms and how they function in PsA transition. This information can further be crucial for treatment protocols in PsA (25).
3.5.3. Metabolomics (Lipidomics)
Specimens like human plasma, urine, and feces, containing abundant metabolites with abnormal disease-specific temporal profiles, are being tested. These include modified levels of glucuronic acid, α-ketoglutaric acid, serum choline metabolite trimethylamine N-oxide (TMAO), and differences in bile acids (glycoursodeoxycholic acid sulfate) and butyrate in PsA development (28). Glycerol 1-hexadecanoate, α/β-turmerone, dihydrosphingosine, glutamine, and pantothenic acid have been elucidated as potential fecal metabolic biomarkers. Urine metabolomics has shown elevated glutamine, histamine, xanthine, phenylacetic acid, xanthurenic acid, lignoceric acid, and creatinine, with reduced citrate, alanine, methylsuccinate, trigonelline, ethanolamine, p-hydroxyphenylpyruvic acid, and phosphocreatine in PsA patients (14, 19, 28).
3.5.4. Single-Cell Phenotyping and Quantitative “Tissue Fluorescence-Activated Cell Sorting Analysis”
Single-cell phenotyping of immune cells and their interaction in PsA pathogenesis has emerged as a diagnostic approach with a key focus on immune checkpoint signaling, as demonstrated through fluorescence-activated cell sorting (FACS) in early studies. These technologies contribute to understanding the cellular mechanisms involved and can assist in developing therapeutic agents targeting those mechanisms (28).
3.5.5. Digital Technology
Digital technology is the latest weapon in the armory for detecting early PsA, aimed at filling the delicate gap between developing PsA and visible symptoms. The digital tools, apart from electronic records and virtual visits, include telemedicine, smartphone technology, artificial intelligence, and machine learning (29, 30). Machine learning algorithms are the newest tool under evaluation for diagnosing early PsA with high accuracy and precision (31, 32). Shapiro et al. devised the ‘PredictAI™’ model to accurately detect undiagnosed PsA (33) Similarly, a specialized ‘Disease Activity Score 28 (DAS28-CRP)’ utilizing machine learning-based permutation targeting ‘tenderness’ in PsA activity staging is also being developed (34). Machine learning analytics have also been expanded to identify distinct PsA phenotype clusters based on joints and skin/nail involvement, dactylitis, enthesitis, and axial manifestations (35). Using unsupervised clustering analysis, the machine learning algorithm ‘FlowSOM’ revealed higher memory T-helper (Th) cells in PsA (36). Some PsA risk prediction models are using 2-digit HLA alleles imputed using the SNP2HLA algorithm for stratifying and validating datasets to effectively segregate at-risk PsA patients (37). Using PsA-specific immune profiles via a machine learning approach, Mulder et al. showed an association with CD197+ monocytes and memory CD8+CD45RACD197- effector T-cells. Along with in-depth flow cytometry, this machine learning could identify distinct PsA immune cell profiles (2). ResNet neural networks aim to detect early PsA based on MRI-dependent joint inflammatory patterns, while the Disease-gene Immune cell Expression (DIME) network captures crucial PsA genes (38, 39).
Digital tools are now a revolution in medicine. The employment of various modes of digital technology can effectively harness multidimensional information, ranging from not just the classical factors like patients’ clinical features, imaging and serological findings, but even more specific genetic, metabolic, and immunologic profiles. The digital models have the advantage of being able to scan and analyze large patient databases for suspected undiagnosed PsA patients. Also, information from massive electronic medical records can be easily and rapidly assessed, thus tackling the manpower logistics (33-35, 39). Validated models can, in the future, serve as important point-of-care support tools for the general population, greatly reducing the need for more elaborate and time-consuming laboratory testing. Flagging even the subtle signs can address the outlier patients, which is currently a challenge for clinicians (35). Appropriately designed machine learning and AI models can provide the benefit of specific focus on relevant factors and can further be tailored to include treatment response as well. These tools can reduce the overall variability in the assessment and classification of at-risk PsO patients and also recognize patterns that may be missed by humans (33-35, 39). Thus, digital technology is crucial for early identification and interception of disease progression in PsO patients.
4. Conclusions
Several clinical trials have emphasized that even a slight delay in rheumatologic intervention in PsA can lead to irreversible joint damage, motor disability, and an overall impaired quality of life (1, 4). Timely and accurate identification of ‘at-risk’ PsO patients will serve as the foundation for appropriate intervention. The important checkpoints for this intervention include the timely detection of early PsA and its differentiation from other common PsA-mimicking arthropathies (osteoarthritis, OA, and fibromyalgia, FBM). An accurate prediction of PsA development will allow for escalation or modification of ongoing treatment, making it more comprehensive, preventing misdiagnosis and overtreatment due to mimickers, and facilitating prognostic and monitoring information (4, 8, 14, 15).
Hence, a holistic multi-specialty approach encompassing multi-omics, machine learning, and artificial intelligence, in addition to conventional investigative modalities, appears extremely promising with its successful utilization. This strongly underlines the need for close collaboration between dermatologists (the first point of contact for PsO patients), immunopathologists (investigative workforce), and rheumatologists (late consultation contact for PsA).