1. Introduction
Skin is the largest body organ in vertebrates and plays a vital role in various functions; it serves as an interface with the external environment, and maintains key homeostatic functions during the lifespan (1, 2). Skin regeneration is a significant domain of research in tissue engineering (TE), particularly for massive skin loss cases, where existing treatments are not yet able to induce permanent complying skin regeneration (3). The regeneration ability of human skin is currently well accepted, but the question for researchers and clinicians is how to restrain this innate potential to treat cutaneous injuries and diseases or disorders. The skin is a highly complicated and dynamic system consisting of numerous cell types and matrix components. Multiple stem cell populations are recognized in skin and these cells act a pivotal role in skin development, repair, and homeostasis. In general, stem cells are defined by their capability to self-renew and their capacity to differentiate into function-specific daughter cells. These progenitor cells are isolated from all skin layers (epidermis, dermis, and hypodermis) and have sole complementary roles in maintaining the skin integrity. The skill to understand and regulate these stem cell populations to promote skin regeneration is where the promise of regenerative medicine lies on (2).
Stem cells are known as basic cells that are primarily unspecialized and can generate several cell lineages (4), with 3 particular characteristics as follows: 1- Self-renewal, which is the most crucial property of a stem cell; i e, division resulting in similar cells; 2- Specialized cells; the ability to regenerate the tissue in which it is located; and 3- Plasticity, which is the ability to generate other cell types different from those of the original tissue, and make stem cells (4, 5). From the potency point of view, they can be classified into 5 groups including totipotent (the zygote), pluripotent (inner cell mass of the blastocyst), multipotent (mesenchymal/hematopoietic stem cells), oligopotent, and unipotent (tissue specific stem cells) (6). On the other hand, they are divided into 2 main groups: embryonic stem cells and adult stem cells available in the blastocyst (the inner cell mass of an embryo 5 days after fertilization of the egg by the sperm) and come from the formation of the fetus, respectively. They are responsible for repairing damaged tissue composed of cells in more differentiated stages (5). Although the adult epidermis basal layer contains stem cells known for years, it is not obvious yet whether all cells are stem cells or just a small number of them exists within this layer. The epidermal proliferative unit (EPU) covered with a hexagonal surface cell, is a bed for 10 firmly packed basal cells yielding a pile of increasingly larger and flatter cells. The hypothesis that there is a self-renewing stem cell per EPU and that the other basal cells are so-called transit-amplifying (TA) is derived from this (7).
A wide variety of environmental factors contribute to the overall control of stem cell behavior (8). Growth factors are one of the factors used to differentiate stem cells in vitro. The ongoing prevalent method to differentiate stem cells is the use of culture medium supplemented with enough amounts of growth factors. But, Brownian motion of growth factors in the medium leads only a small amount of them to reach biological signal pathway cell receptors. Hence, even if a large amount of growth factors is added to culture medium, only a small portion is involved in the differentiation of stem cells (9). Moreover, stem cells can be stimulated by chemical or topographical cues (10). At the moment, it is believed that stem cells can sense and initiate an appropriate response to the physicochemical properties of their extracellular matrix (ECM) or any substrate that they are cultured on, by regulating their complex signaling pathways (11-16). Cells are naturally located in a chemically and topographically complicated environment in vivo, which is different from what is used commonly for in vitro culture conditions (17-19). Several sizes of topographies, from macro to micro, and nano scale featured cells might be faced (20). In response to topographical cues tested by nano scale patterns such as dots, columns, pores, pits, meshwork, gratings, and random surface roughness, cells undergo different reactions in terms of morphology, adhesion, proliferation, cytoskeletal formation, migration, gene expression, and surface antigen display. The previously mentioned patterns are created by a variety of techniques such as nanolithography and nanofabrication or molding (8, 10, 11, 20-22).
2. Methods
2.1. Human Primary Keratinocyte Isolation and Culture
2.1.1. Sample Preparation
Foreskin tissue was taken from a newborn baby during circumcision with the informed consent from their parents and kept in cold EpiLife (Gibco) containing 300 U/mL penicillin and 300 µg/mL streptomycin (Gibco) at 4°C until use. The tissue sample was placed in an uncoated 100-mm bacterial Petri dish and kept moist with some medium. Subcutaneous fat and loose connective tissues (hypodermis) were removed using fine tweezers and a scalpel.
2.2. Overnight Digestion
Then, the skin was placed in a Petri dish, while the epidermis side was down. Tissue was scraped away using the edge of the scalpel until only the thin epidermis and the dense dermis remained, and cut the piece into ~3 - 4-mm wide strips. Pieces were placed with the dermal side down in a 60-mm dish containing 5 - 6 mL of EpiLife containing 300 U/mL penicillin and 300 µg/mL streptomycin (Gibco) and Dispase. Then, covered and kept in a sterile place at 4°C for 12 - 16 hours (overnight).
2.3. Epidermis Isolation
The overnight digested tissue pieces were taken and the epidermis was slowly peeled off from dermis using a tweezer grabbing the edge of the dermal part of the tissue and the thin epidermal part with another set of thin tweezers. The epidermis (almost transparent) was immediately transferred into another dish with EpiLife medium and cut into small pieces of 1 - 2-mm width using scalpel and transferred to a falcon tube containing 5 mL of TrypLETM Select (Gibco). Samples were incubated at 37°C for 20 - 25 minutes and mixed gently every 5 minutes until the solution became turbid. About 5 mL of the medium (e g, DMEM) containing a minimum of 10% FBS was added to the solution and pipetted vigorously up and down for about 10 - 15 times. Undigested pieces of tissue were eliminated by passing the solution through a 70-µm mesh filter into a new Falcon tube. The solution was centrifuged at 200 g for 5 minutes and the supernatant was discarded and the cell pellet was resuspended in 5 mL EpiLife medium. Number of cells was counted (e g, with a hemocytometer).
2.4. Keratinocyte Culture
For a better result, culture plates were coated with Coating Matrix (type I collagen) by adding ~2 mL coating solution to each well of 6-well tissue culture plates and incubated for at least 30 minutes at 37°C. The coated dishes were stored at 4°C for several days. Coating Matrix was removed and seeded out about 4 × 104 cells per cm2 in an appropriate EpiLife medium containing 5% HKGS (human keratinocyte growth supplement) and incubated at 37°C, 5% CO2 and 95% humidity. Medium was changed the next day and subsequently every 3 days. When cells reached 70% - 75% confluency, they were rinsed with phosphate-buffered saline (PBS) (without calcium), trypsinized using TrypLE Select, and incubated at 37°C. Cells were rounded up and came off the plate within 4 - 5 minutes.
2.5. Keratinocyte Fixation and Substrate Development
The cell medium was completely removed from the plates before the next step. Cells were fixed with 4% paraformaldehyde to maintain their shape before molding process. Polydimethylsiloxane (PDMS, SYLGARD® 184, RTV, Dow Corning, USA) was used to mold and fabricate the desired substrates. For this purpose, the silicone resin and curing agent were mixed in 10:1 ratio, and then, heated for 30 - 35 minutes at 45°C by microplate heater. After cooling down the preheated solution to 37°C, the cured silicone was poured onto each of the fixed samples and incubated at room temperature (RT) for at least 24 - 48 hours to obtain the imprinted substrates. Then, the cured silicone was peeled off from cell culture plates. In order to remove cell debris, the substrates were washed with boiled water and 1 M NaOH solution. Another substrate was developed with an empty plate without any fixed cells as a negative control.
2.6. SEM Analysis of the Developed Substrate
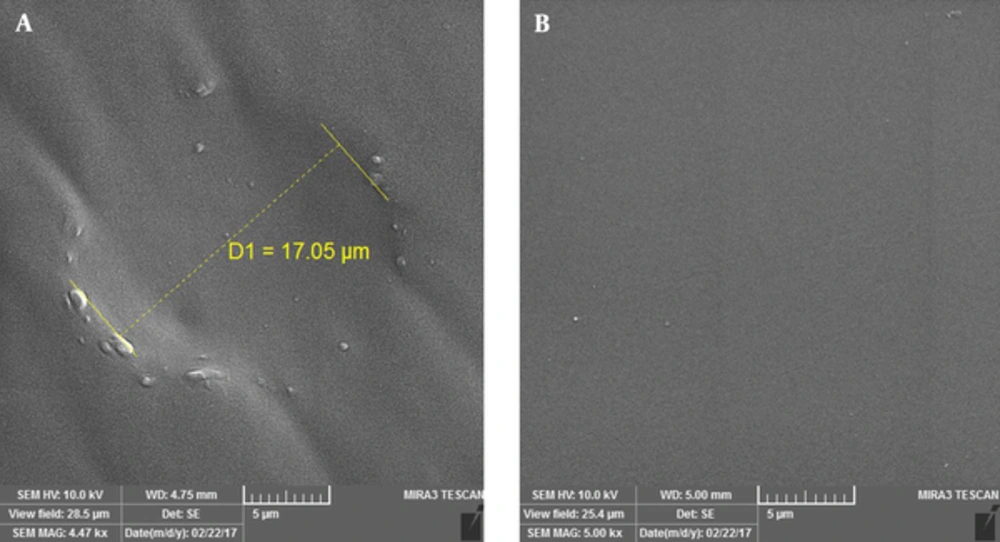
Morphology of the developed PDMS substrate containing keratinocyte shape and the negative control substrate without any cell shape was observed by scanning electron microscope (SEM).
3. Results
3.1. Primary Human Keratinocyte Isolation
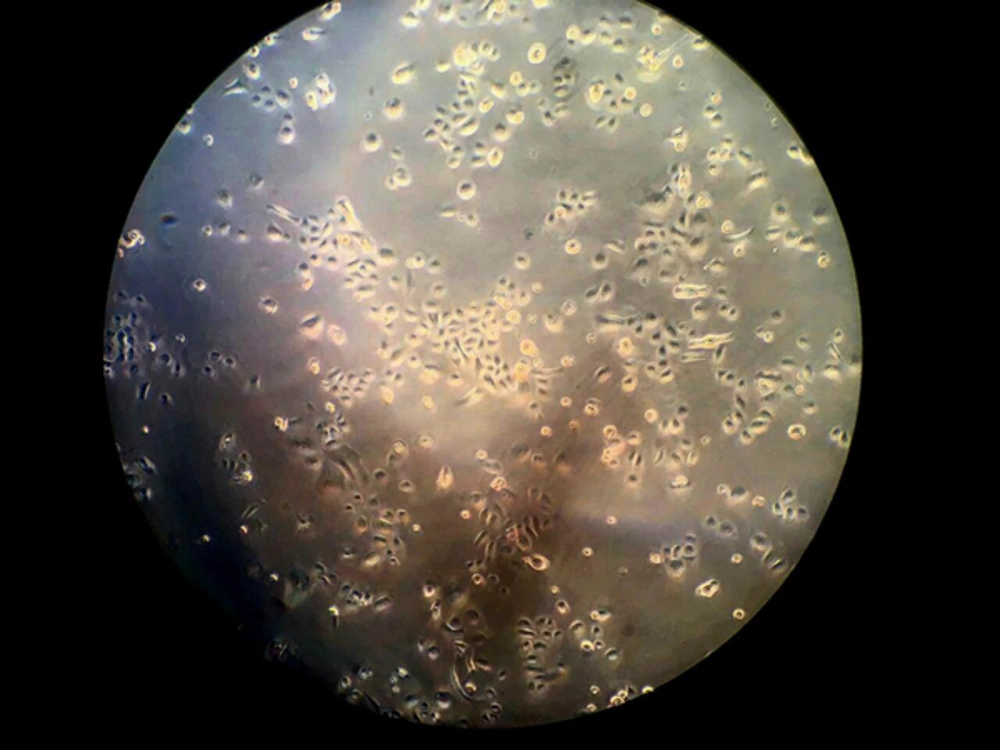
Human primary keratinocytes were successfully isolated from foreskin samples of newborn babies using the previously mentioned method after taking an informed consent from their parents (Figure 1). Isolated keratinocytes showed a successful culture in EpiLife medium containing 5% HKGS plus 300 U/mL penicillin and 300 µg/mL streptomycin. Collagen type I also helped better attachment of the isolated cells. As shown in Figure 1, cells were observed using an invert microscope (Figure 2).
3.2. PDMS Substrate Development and Analysis
Keratinocytes were fixed by paraformaldehyde solution and used for PDMS substrate development for cell shape molding. As previously mentioned, the silicone was cured and poured on the fixed cells and the desired substrate was successfully developed. SEM imaging analysis confirmed the formation of the cell shape grooves on PDMS silicon as shown in Figure 3.
4. Discussion and Conclusion
Skin regeneration is one of the most important fields in the regenerative medicine. There are different types of stem cells, which are essential in regenerative medicine and tissue repair. Each one has its own advantages and disadvantages by which they can be selected for various medical and research applications. They play a crucial role in all fields of regenerative medicine due to their huge capacity to differentiate and produce different kinds of specialized cells related to the repair of the damaged tissue (3, 5-7). When culturing stem cells in vitro, it is important to provide normal conditions for cells, which is essential for their optimum propagation and differentiation. One of the essential conditions is the need for a well-developed environment, which resembles their natural ECM (2, 8, 11). For this purpose, researchers used different topography shapes, which can affect the stem cell differentiation. Previous researches showed that different topographies with various shapes (i e, triangle, round, multigonal, etc.) or sizes (from micro to nano) can affect and lead stem cells differentiate into the desired specialized cells (8, 11, 20-22). Based on this idea, the current study developed a substrate containing the keratinocyte shape printed as topography on the PDMS silicone and aimed using it as a culture substrate for further stem cell culture and stem cell differentiation studies. Therefore, in addition to keratinocytes, other types of cells can be utilized to develop such topographies and different stem cells can be cultured on the developed substrates.