1. Background
Endocarditis is a rare infectious disease of the cardiac endothelium. The incidence rate ranges from 1.5 cases to 9.6 cases per 100,000 people, but the mortality rate is nearly high (about 25%) (1). Usually, a new cardiac problem with symptoms of high fever, sepsis, and systemic problems represents the likelihood of infective endocarditis (2). Symptoms are often ambiguous, so recognition is sometimes delayed, which can affect the quality of life (3). Early detection of infective endocarditis is critical, as treatment should be administered at a specific time to achieve the correct outcome (1). Different laboratory techniques such as culturing, immunohistochemistry and immunofluorescence tests, serology, and molecular techniques are used for the diagnosis of endocarditis. Among the identification methods, isolation of bacterial strains by the culture of cardiac vegetation is a relatively available method to detect infective endocarditis. However, the negative result of culture does not rule out endocarditis (4). In other words, blood culture-negative endocarditis (BCNE) should be considered as a possible cause and contestable matter. Antibiotic use, bacterial growth conditions, and immunocompromised status are some of the BCNE etiologies. Mitral and aortic valves are most commonly affected, and tricuspid endocarditis has been reported in a few studies (5). Different microorganisms which cause BCNE include Coxiella burnetii, Bartonella species, Mycobacteria, and Tropheryma whippelii (6).
Using only culture-dependent methods, as is common in most diagnostic centers, could be unreliable. Therefore, to obtain reliable results, molecular methods, including real-time PCR, should be performed. In this way, fewer false-negative results are obtained (7). The mortality rate of 20% in endocarditis and the severity of the disease may be because treatment could depend heavily on the detection of the microorganisms. Therefore, the use of an appropriate detection method is critical (8). Recent studies of patients with overt endocarditis symptoms who have a negative cultural and serologic test and a positive 16S ribosomal ribonucleic acid (16SrRNA) result indicate that molecular techniques are the best method for detecting BCNE (9). The state of immunosuppression could affect the serological result and negative results may mislead the physician to recognize the disease and apply the appropriate treatment (10). In addition to increasing biosafety and reducing the subculture of microorganisms, the use of PCR testing could increase the rate of confirmed positive BCNE tests up to 24.3% (11). There are a few studies about the rate of BCNE in Iran, which reported high rates, about 35% (12). In recent years, most investigations of endocarditis in Iran have used methods other than molecular testing to detect endocarditis. In 2010, the use of serological and microbiological tests showed that Brucella spp. was isolated in 2% of endocarditis cases (13). Another study in 2017 also showed a high prevalence of blood-negative culture endocarditis based on microbiological tests (14). However, in 2018, molecular tests showed the presence of some difficult-to-culture-bacteria including, C. burnetii (30.77%) (15) and as mentioned above, the importance of using molecular methods should be considered a definite detection method.
2. Objectives
This study aims to assess the prevalence of possible infectious agents causing endocarditis in patients who had heart surgery and negative culture history.
3. Methods
3.1. Patients and Histopathology
Samples selected in this investigation referred from 2012 up to 2018 to Rajaie Cardiovascular Medical and Research Center as one of the referral centers in Iran which were reserved in paraffin-embedded tissue collection of this center. Including criteria for selection was the presence of inflammation in pathologic investigations based on the modified Duke criteria and negative results in blood culture tests. All parts of the sample selection in this study were approved by the Ethics Committee of the Pasteur Institute of Iran (Code: IR.PII.REC.1399.051).
3.2. DNA Extraction from Paraffin-Embedded Tissue Samples
To extract DNA from paraffin-embedded samples, tissue cores or microdissected tissue was cut into small parts and then subjected to xylene treatment, which dissolves the paraffin from the tissue, and then rehydrated using a series of ethanol washes (16). Genomic DNA was extracted from proceed samples using QIAGEN DNeasy® Blood & Tissue (Qiagen, Germany), according to the manufacturer’s instructions. All extracted DNA was stored at -20 °C until molecular testing.
3.3. Molecular Identification of Bacterial Pathogens
Different molecular techniques were used to identify bacterial agents. Besides the real-time PCR method, for detecting Coxiella burnetii, Tropheryma whipplei, and Brucella spp., PCR sequencing was performed to identify Legionella spp. and Bartonella spp. since detecting the exact species of the mentioned bacteria was important in the current study. Also, for the samples with negative results of PCR sequencing and real-time PCR, 16srRNA PCR was used to determine the bacterial causative agent. The list of primers is available in Table 1.
| Gene | Forward Primer | Reverse Primer | Probe |
|---|---|---|---|
| IS1111 (Coxiella) | AAAACGGATAAAAAGAGTCTGTGGTT | CCACACAAGCGCGATTCAT | 6-FAM-5'-AAAGCACTCATTGAGCGCCGCG-3'TAMRA |
| IS711 (Brucella spp.) | GCTTGAAGCTTGCGGACAGT | GGCCTACCGCTGCGAAT | 6-FAM-5'-AAGCCAACACCCGGCCATTATGGT-3'TAMRA |
| WISP family protein (Tropheryma whipplei) | TTGTGTATTTGGTATTAGATGAAACAG | CCCTACAATATGAAACAGCCTTTG | 6-FAM-5'-GGGATAGAGCAGGAGGTGTCTGTCTGG-3'BHQ-1 |
| 16S rRNA gene (Legionella) | AAGATTAGCCTGCGTCCGAT | GAGGGTTGATAGGTTAAGAGC | - |
| Bartonella 16srRNA | TTAGAGTGAGCGGCAAAC | TACCGTCATTATCTTCACCG | - |
| 16S rRNA | AGAGTTTGATCMTGGCTCAG | AAGGAGGTGWTCCARCC | - |
Primer Sequences Used in This Study
3.4. PCR Sequencing Assay for Detection of Legionella spp. and Bartonella spp.
PCR tests were performed to detect Legionella (23S-5S rRNA gene) and Bartonella spp. The optimized PCR reaction conditions were 95°C for 60 s, followed by 40 cycles of 95°C for 30 s, 50°C (Legionella) and 60°C (Bartonella) for 30 s, and 72°C for 30 s. The list of primers is available in Table 1. All the tests have been done by Labcycler Gradient SenSoQuest (Yechizi Co, Germany). Specific positive controls were used in all tests. Following gel electrophoresis, all positive samples were sequenced.
3.5. Real-time PCR Assay for Detection of Coxiella burnetii, Tropheryma whipplei, and Brucella spp.
The bacterial genera, including C. burnetii, T. whipplei, and Brucella were investigated through real-time PCR. Reactions were prepared using the following mixture: 12.5 μL of 2x Master Mix (RealQ Plus for Probe, Ampliqon, Denmark), 900 nM from each primer, 200 nM probe, and 5 μL of DNA templates. The Corbett 6000 Rotor-Gene system (Corbett, Victoria, Australia) was used for performing the tests, with a final volume of 25 µL. The PCR amplification program was 10 mines at 95°C, followed by 45 cycles of 15 s at 94°C and 60 s at 60°c.
3.6. 16srRNA Sequencing
PCR test followed by sequencing was used to detect the 16srRNA gene to determine the causative bacterial agent. The optimized PCR reaction conditions were 95°C for 60 s, followed by 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. All the tests have been done by Labcycler Gradient SenSoQuest GMBH. Specific positive controls were used in all tests. All positive samples were sequenced. Sequenced on both strands using the BigDye Terminator v. 1.1 Cycle Sequencing Kit® (Applied Biosystems) and an ABI PRISM 3130xl Sequencer® (Applied Biosystems) were sequenced. The sequences thus obtained were compared with the sequences available in GenBank using the BLASTN program (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Species identification was based on 99% 16S rDNA sequence identity with the GenBank prototype sequence, and genus identification was based on 97%.
4. Results
4.1. Patients and Sampling
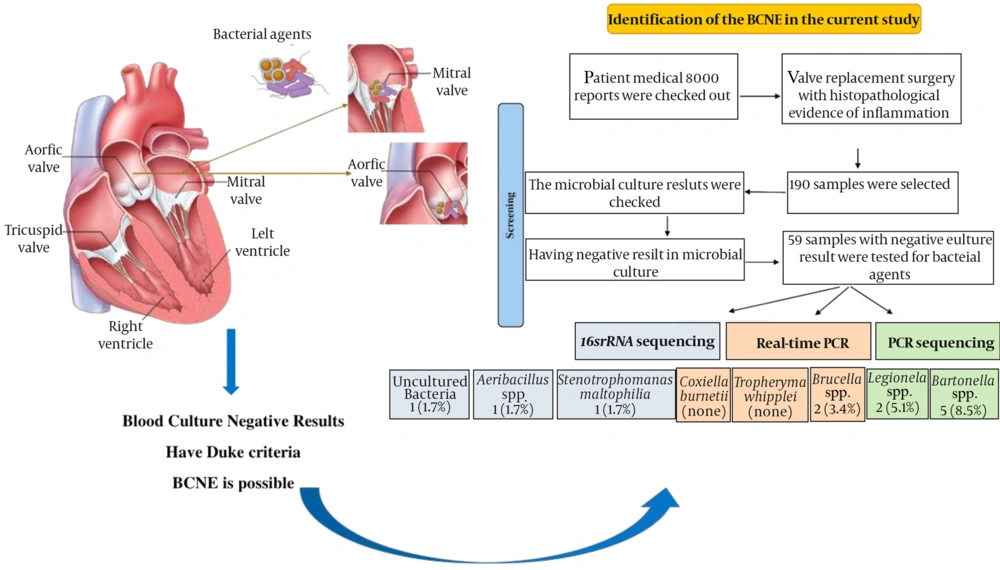
Over the study period, eight thousand patient medical reports between 2012 to 2018 were checked out, and 190 samples were selected from patients who had valve replacement surgery with histopathological evidence of inflammation and according to the modified Duke criteria. Out of 190 samples, 59 specimens had a culture-negative result and were without any evidence of confirmatory data on a causative agent. 18 (26.4%), 21 (30.8%), 4 (5.8%), 5 (7.3%) were categorized into sub-acute, chronic inflammation, acute inflammation, and acute-chronic inflammation, respectively. The rest belonged to other pathological reports without considering the inflammation or bacterial presence (Figure 1).
4.2. Molecular Detection of Bacterial Agents
4.2.1. Results of PCR Sequencing Assay
The PCR sequencing results indicated that among 59 samples with negative culture results, Bartonella and Legionella were detected in 5 (8.5%) and 3 (5.1%) samples, respectively. According to sequencing results, all samples of Legionella were L. steelei and all the Bartonella samples were Bartonella quintana.
4.2.2. Results of Real-time PCR Assay
According to the results of the real-time PCR assay, C. burnetii and T. whipplei were not detected while Brucella was detected in 2 (3.4%) samples.
4.2.3. Results of 16srRNA Sequencing
16srRNA sequencing assay revealed that there were seven samples with positive results and the sequencing showed one Stenotrophomonas maltophilia, one Aeribacillus spp., and one uncultured bacterium.
5. Discussion
BCNE could be considered as one of the contestable diseases since the diagnosis is usually associated with some difficulties. The use of antibiotics and the presence of fastidious bacteria and intracellular microorganisms that cannot be detected in blood culture tests are some of the reasons that complicate the diagnosis. The rate of BCNE is reported to be about 35% (7), however, this rate varies from 10.2% to 75.6% in different countries because of methods and strategies for detection, rates of fastidious microorganisms, history of antibiotic use specifically before blood collection, methods of sampling, and the presence of noninfectious or unknown pathogens may vary in different regions (17). In addition, the rate of IE is considerable in southern Mediterranean countries, and most patients with IE were younger than 40 years of age (18). The College Center of Aix, one of the valid BCNE detection centers in France, reported 1500 endocarditis with negative culture results from 2010 to 2015 (12). The use of different identification methods in this center could increase the probability of diagnosis. In other words, they indicate that, in addition to serological methods, PCR is a reliable method for detecting challenging microbial BCNE pathogens.
In the current study, 8000 patient medical reports were reviewed, and 190 samples were collected. Of the 190 samples, 59 samples tested negative for bacterial pathogens with culture results. Bartonella, Legionella, and Brucella were detected in 5 (8.5%), 3 (5.1%), and 2 (3.4%) specimens, respectively. The rates of Bartonella and Legionella are remarkable in the current study. Similar results are shown in other studies. Pecoraro et al. reported Bartonella as the most common cause of endocarditis (19). On the other hand, the results showed that 16srRNA sequencing is not a helpful method to detect the different causative agents of BCNE, since it was only detected in 7 samples. Among these seven samples, one Stenotrophomonas maltophilia, and one Aeribacillus isolate were detected.
In the current study, the rate of Bartonella was 8.5%. Bartonella spp. was first discovered as endocarditis causative agent in 1993 (20). Up to 2006, 120 Bartonella spp. were detected as BCNE causative agents (21). Notably, Bartonella is the cause of nearly 12% to 60% of BCNE cases, making it the second most common cause of BCNE after Coxiella burneti (22). Bartonella quintana and B. henselae were the most prevalent agents (23). In the current study, we used two different methods (using species-specific primers and sequencing) to increase specificity. Using an appropriate method to detect bacteria is critical to reduce the number of false positives and negatives. For example, serologic tests are useful laboratory methods for detecting Bartonella in endocarditis. However, the incidence of cross-reactivity between Bartonella and some other bacteria such as Chlamydia and C. burnetii is problematic. Based on various studies, the tests from RT-PCR are useful, sensitive, specific, and rapid methods for the detection of Bartonella in endocarditis. Real Time-PCR is a specific test for the detection of Bartonella in heart valve samples (92%) (24); while for the blood or serum samples, RT-PCR is less specific (33% and 36%, respectively) (25). Compared with 16srRNA, real-time PCR is a more sensitive test; however, all positive real-time PCR tests also had positive results for 16srRNA sequencing in the current study. On the other hand, endocarditis caused by Bartonella occurs more frequently in patients with valvular insufficiency and valvular heart surgery, which is why detection of Bartonella at the right time would be so important. In the future, the development of PCR methods to detect Bartonella could be a rapid, reliable, and preventive test to detect endocarditis in our hospitals.
In the present study, Legionella was detected in 5.1% of the samples with negative culture results. Endocarditis caused by Legionella has rarely been reported. It was first discovered in 1984, and only 16 cases had been reported by 2016. Of these 16 patients, only two had healthy and normal heart valves, and the others had a history of valve surgery (26). PCR and culture-based methods are the gold standards for Legionella detection. In addition to these gold-standard methods, the detection of bacterial antigens in urine samples is also a routine method (27), however, the delay in the increase of antibodies remains a significant problem. In other words, a positive result on pathologic testing, observation of bacteria through an electronic microscope, and positive PCR results are the accurate ways to detect Legionella in endocarditis (28). As far as we know, this is the first report of the detection of Legionella species other than L. pneumophila causing BCNE in Iran.
In this study, Brucella was detected in 3.4% of BCNE cases. Endocarditis caused by Brucella is endemic in some regions such as Egypt, and it is usually a common causative agent of BCNE, after Bartonella (8). Although Brucella was less common, Brucella melitensis has been reported to be associated with a history of valvular heart surgery, which can cause more severe disease. In other words, without effective and timely antimicrobial treatment and valve surgery, the mortality rate could be as high as 80% (29). Iran is an endemic region for brucellosis, and the Ministry of Health and Medical Education has classified the country into four areas: very high, high, average, and low prevalence. For this reason, it seems that endocarditis may have a high prevalence in our country. Misdiagnosis is common because brucellosis has no pathognomonic symptoms. Therefore, the use of a rapid and sensitive method is crucial.
We also found Stenotrophomonas maltophilia among the BCNE samples. Stenotrophomonas forms a biofilm that can adhere to various surfaces and has a high resistance to antimicrobial agents. S. maltophilia has a low probability of causing endocarditis as a nosocomial pathogen. Although it could be considered a multidrug-resistant isolate, Stenotrophomonas is an opportunistic bacterium and should be considered in patients with underlying diseases (30). In other words, intrinsic resistance leads to an increase in morbidity and mortality among BCNE cases; therefore, timely identification of this organism is necessary to improve medical management. It seems that the development of real-time PCR-based methods may have advantages in reducing mortality and morbidity rates in the future.
5.1. Conclusions
According to the current results, the factors causing endocarditis could be different. The rate of Legionella and Bartonella quintana, which are difficult to detect by routine laboratory tests, was high in our study, and to the best of our knowledge, this is the first report of Legionella steeli and Bartonella quintana among BCNE cases in Iran. It seems that hospitals should use special technology to detect these BCNE pathogens. In this case, surgery and valve replacement could be avoided. Systematic examination of samples of replaced heart valves to detect fastidious bacteria in cardiovascular centers, consideration of the bacterial group HACEK, and timely detection of BCNE pathogens could prevent heart valve surgery and improve the success of antimicrobial treatment.