1. Background
Echinococcosis/hydatidosis is caused by the larval stages of genus Echinococcus. It can be seen in two major forms: Cystic Echinococcosis (CE) and Alveolar Echinococcosis (AE). These forms of the disease are reported from some countries in the Middle East; however, CE has been much more abundant and more important in this region (1). In addition, CE is one of the most important zoonotic parasitic diseases worldwide (2). The cycle of the parasite is maintained by herbivores including sheep, cows, camels, goats, pork, horses, donkeys, and some wild ones as intermediate hosts and carnivores of Canidae family as definite hosts (3).
The definitive diagnosis for most cases of Cystic Echinococcosis in man is possible by physical imaging methods, such as radiology, ultrasonography, computed axial tomography, and magnetic resonance imaging (4). Serological methods such as ELISA, western blot, IFA, IHA, and CIEP are the most common immunological methods for the CE diagnosis (5). Several drugs have been investigated for the treatment of human CE (6, 7). However, it seems surgery remains the mainstay of therapy for large cysts, superficial and infected cysts, those in vital anatomical locations, and those entering substantial mass effect (8). However, this method has many costs and risks including the probability of shock during surgery, cyst rupturing and expanding to other parts of the body (8).
With the development of DNA-based molecular techniques, huge changes occurred in the phylogenetic classification of different living organisms. Some molecular methods have been used to reveal the intra-species variations among different isolates of Echinococcus granulosus including: randomly amplified polymorphic DNA polymerase chain reaction (RAPD-PCR) (9, 10), restriction fragment length polymorphism (RFLP) (11, 12), single strand conformational polymorphism (SSCP) (13), and sequencing. Sequencing has shown the highest discriminatory power and it now is routinely used for genotyping (14, 15).
By using DNA based molecular techniques, ten genotypes (Gs) with noticeable host specificity have been described in E. granulosus species, most of which have been considered as the causative agents of CE in man. These genotypes include G1-G3 (E. granulosus sensu stricto), G4 (E. equines), G5 (E. ortleppi), G6 - G8, and G10 (E. canadensis) (3). Among them, G1, G6, and recently G2 and G3 genotypes have been found in human and animal reservoirs in Iran using different genetic markers (16-19). However, there has been no study on the genotyping of hydatid cysts in South Khorasan province, Eastern Iran.
In the present study, to clarify the population structure and phylogenetic relationship of CE agents in Eastern Iran, samples of patients undergoing surgery at Birjand hospitals in a ten-year period were studied by sequence analysis of mitochondrial genes of NADH dehydrogenase subunit 1 (nad1) and cytochrome oxidase c subunit 1 (cox1).
Despite several studies have been conducted to determine genotypes of E. granulosus in human and animal hosts in different parts of the world, little information has been collected about the relationship between parasite genotypes and their pathogenesis in the hosts. In this study, along with the determination of genotypes, some phenotypic traits of patients are also considered.
2. Methods
2.1. Samples
In this cross-sectional study, nine hydatid cysts surgically removed from patients with CE were studied. These patients were operated between 2006 and 2015 at Imam Reza hospital in Birjand, Eastern Iran. The diagnoses were confirmed retrospectively by pathologists from the department of pathology, Birjand University of Medical Sciences. The samples included 4 paraffin-embedded CE samples, 3 haematoxylin-eosin (HE)-stained sections, and 2 CE cysts, which surgically were removed during the study period. The profile of the cyst size, presence/absence of protoscoleces, history of previous surgery, and cyst location in each patient were considered.
2.2. DNA Extraction
The paraffin-embedded tissue blocks were cut into sections with a thickness of 5 - 10 µm. For H and E stained tissue smears, the material was scraped using a sterile scalpel. The tissue sections and scrapings were deparaffinized using xylene. Then, the re-hydration process was done in 100%, 90%, 80%, and 70% ethanol (4). For ethanol preserved samples, a germinal layer of the cysts was separated by a scalpel. Then, DNA was extracted using Dynabio Tissue DNA Extraction Mini Kit (Takapouzist, Iran) according to the manufacturer’s instructions.
2.3. Polymerase Chain Reaction
The Polymerase chain reaction (PCR) amplification of the mitochondrial NADH subunit I (nad1) gene was performed by using MS1 (5’-CGTAGGTATGTTGGTTTGTTTGGT-3’) and MS2 (5’-CCATAATCAAATGGCGTACGAT-3’) primers (17), which could synthesize a 397 bp long fragment. For the amplification of the mitochondrial cytochrome c oxidase subunit 1 (cox1) gene, the primer pairs JB3 (5’-TTTTTTGGGCATCCTGAGGTTTAT-3’) and JB4.5 (5’-TAAAGAAAGAACATAATGAAAATG-3’) (20) were used to amplify a 420 bp fragment.
The PCR amplifications were performed in a Mastercycler gradient thermal cycler (Eppendorf, Germany). Positive and negative controls were normally run with every PCR master mix. The PCR products were electrophoresed on agarose gel (1.7%), stained with ethidium bromide, and visualized by a transilluminator. As the size reference, a 100 bp ladder (Jena Bioscience, Germany) was used.
2.4. DNA Sequencing
The PCR fragments were submitted to Bioneer Company (South Korea) and subjected to sequencing by an applied biosystem automated sequencer (3730 XL). Sequencing was performed in both directions using previously mentioned PCR primers using the BigDye Terminator v3.1 cycle sequencing kit.
2.5. Phylogenetic Analysis
Phylogenetic trees were generated by using representatives of Birjand sequences, as well as available E. granulosus G1 and E. canadensis G6 sequences from other regions of Iran and the most relevant sequences isolated from other parts of the world (Table 1). Sequences were aligned in ClustalW and the alignment was manually refined by BioEdit software (version 7.2.5). The non-repetitive region was used for tree construction. Maximum likelihood (ML) trees were inferred by MEGA 6 software (21). Nodal support was assessed by bootstrapping with 1000 replicates. The Ethics Committee of Birjand University of Medical Sciences approved the ethical considerations of the present study under No Ir.bums.REC.1394.421
| Accession Numbers | Genotype | Host | Location | References |
|---|---|---|---|---|
| AB813183 | G6/7 | Wolf | Mongolia | Yanagida et al. (2013) |
| AB813182 | G6/7 | Wolf | Mongolia | Yanagida et al. (2013) |
| AB893252 | G6/7 | Human | Mongolia | Ito et al. (2014) |
| AB893255 | G6/7 | Human | Mongolia | Ito et al. (2014) |
| AB893259 | G6/7 | Human | Mongolia | Ito et al. (2014) |
| AB893258 | G6/7 | Human | Mongolia | Ito et al. (2014) |
| AB893263 | G6/7 | Human | Mongolia | Ito et al. (2014) |
| AB777922 | G6 | Camel | Ethiopia | Nakao et al. (2013) |
| AB777923 | G6 | Camel | Ethiopia | Nakao et al. (2013) |
| AB777909 | G6 | Wolf | Russia | Nakao et al. (2013) |
| AB688142 | G6 | Human | Russia | Konyaev et al. (2012) |
| KR337812 | G6 | Human | Iran, Birjand | This study |
| KR337817 | G1 | Human | Iran, Birjand | This study |
| JQ250809 | G1 | Human | Iran, Shiraz | Sadjjadi et al. (2012) |
| KX227128 | G1 | Human | Chile | Alvarez Rojas et al. (2017) |
| KX020374 | G1 | Cattle | Armenia | Ebi et al. (2016) |
| AB688140 | G1 | Human | Russia | Konyaev et al. (2012) |
| AB688600 | G1 | Sheep | Jordan | Yanagida et al. (2012) |
| AB787548 | G1 | Human | Mongolia | Narankhajid et al. (2013) |
| KR337820 | G1 | Goat | Iran, Gachsaran | Karamian et al. (2015) |
3. Results
Isolated genomic DNA of hydatid cysts of 9 human cases was tested and nad1 and cox1 loci were amplified from all the purified DNA samples. The sequences of partial nad1 and cox1 genes of these isolates were deposited in the GenBank database under the accession numbers KR337812- KR337818 and KP751432- KP751436.
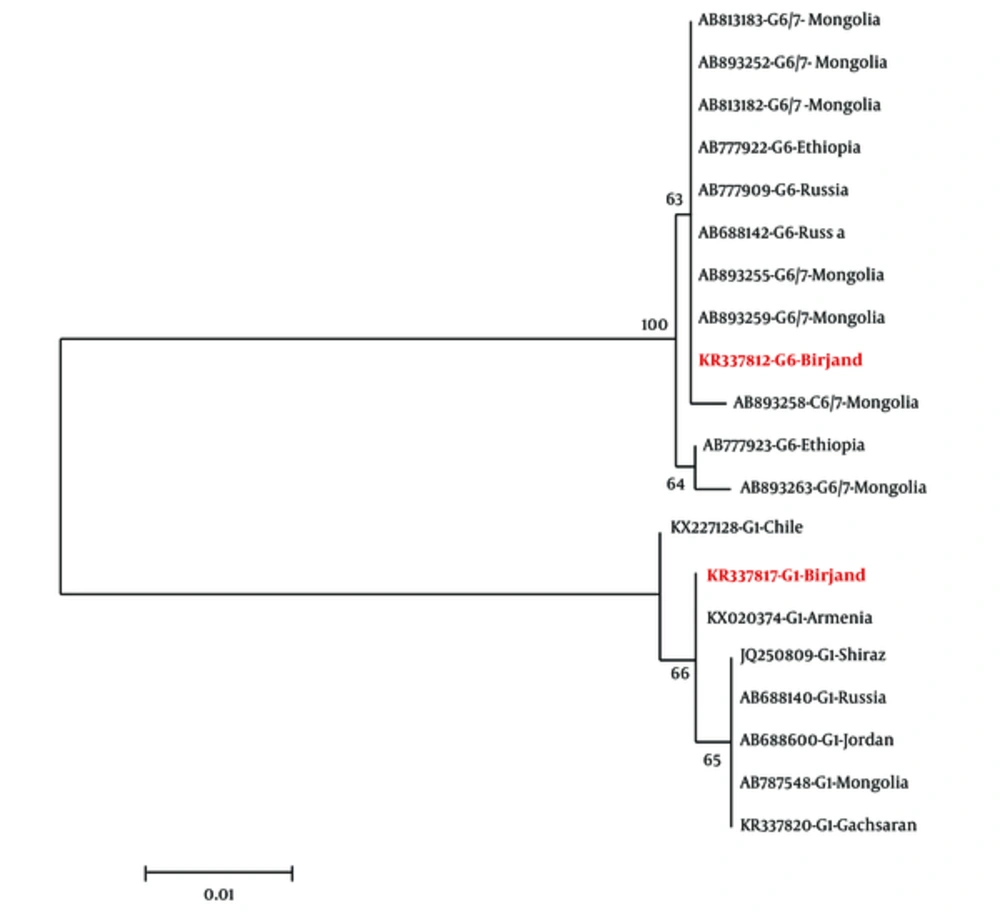
The analysis of cox1 and nad1 gene sequences obtained from the hydatid cysts revealed the dominance of G6 genotype in South Khorasan province, in which eight out of nine cysts belonged to G6 genotype while G1 genotype was found only in one sample. The maximum likelihood tree topology of cox1 region showed all G6 sequences from human cases in East Iran had the maximum similarity with G6 sequences from Mongolia and Russia, Ethiopia, Sudan, and Brazil. However, they were less similar to E. canadensis isolated from other parts of Iran (Figure 1). All the 9 isolates were also genotyped by amplification and sequence analysis of nad1 fragment and the results were consistent with those obtained by cox1sequencing.
With regard to cyst specifications, there are differences among the patients; the volumes of cysts in patients were in the range of 15 mL to 500 mL; the largest cyst with a diameter of 14 cm was observed in the patients. In the patients infected with the G6 genotype of E. canadensis, the primary organ affected was the liver in 4, lung in 3, and peritoneum in one patient (Table 2). Only the patient infected with the G1 genotype of E. granulosusone had a history of hydatid cyst surgery. In this patient, the infected cyst had been removed from the peritoneum.
Among eight human cases infected with the G6 genotype of E. granulosus, seven patients residing in urban or rural areas of Birjand, while the other patient was a resident of Zirkooh in the northeast of South Khorasan province. The hydatid cyst isolated from the peritoneum of the patient who infected with G1 genotype was fertile (containing protoscoleces), while in the patients infected with the G6 genotype of E. canadensis, the overall fertility rate of hydatid cysts was 37.5%.
| Genotype | Parasitized Organ | Number of Cases | History of Previous Surgery | Diameter(Average), cm | Fertility, Percent |
|---|---|---|---|---|---|
| G1 | Peritoneum | 1 | + | 6 | 100 |
| G6 | Liver | 4 | - | 4 | 25 |
| Lung | 3 | - | 7.5 | 33.3 | |
| Peritoneum | 1 | - | 3 | 100 |
4. Discussion
The present study has identified a predominance of G6 genotype of E. canadensis in the study area. The only human case infected with the G1 genotype of E. granolosus was operated in 2005, who was from the rural areas of Sarbisheh in Southeast of Birjand. Since then, all surgically treated patients were infected with the G6 genotype of E. canadensis, all of which (except one) were inhabitants of Birjand or its neighboring villages.
Hydatidosis is endemic in many parts of Iran and imposes a significant economic burden on health care systems in the country. The overall annual cost of this disease in Iran has been estimated at US$232.25 million (22). Based on the studies, genotypes of E. granulosus could create different pathogenesis in human (23, 24). For example, human cysts of G8 genotype in North America often do not cause any clinical symptoms in their host and they usually are characterized when screening for other diseases. These cysts have a slow growth in their hosts, so the researchers have recommended careful medical management rather than aggressive surgery (25). Given the differences in the life cycle and pathogenicity of these genotypes, the appropriate health care and prevention programs should be applied to each one.
G1 is the most frequent genotype in CE cases in the world and G6 and G7 are considered in the next ranks, in sequence (26). This genotype has been considered as the main causative agent of echinococcosis (CE) in the main foci of Iran where sheep and dog have been introduced as the main hosts of the parasite. A higher prevalence of this genotype (sheep strain) may be due to the higher number and diversity of its intermediate hosts. Also, the dominance of G6 in countries such Sudan and Egypt was explained by the lack of suitable intermediate hosts for sheep strain in these regions (27). Although South Khorasan province (East focus of CE) has been considered as one of the most important centers for camel breeding (30000 camels in 2014), the number of sheep and goats was prominently higher (about 1,000,000) in this region (28). Sheep are rarely infected by G6 genotype and the consequent cyst is free of protoscoleces. Therefore, the frequency of intermediate host cannot explain the dominance of G6 in this region. The arid and warm weather and vast deserts in Eastern Iran, which is similar to countries such Sudan, Mauritania, and Egypt where G6 is dominant in human cases, may explain the effect of environmental conditions on the cycle of the disease. Molecular research showed the important role of G6 genotype in the cycle of disease in Iran where the increasing number of human infection with G6 has been reported in recent years. While the incidence of CE caused by G6 was 9.1% in 2002, a study in 2015 revealed that 40.8% of CE patients in Central and Southeast regions of Iran were infected by G6 genotype (19). The highest incidence of CE due to G6 was now reported in Eastern Iran where 8 out of 9 human cases (more than 88%) were infected by G6 genotype. Accordingly, the extensive drought in different regions of Iran in recent decades (19) can be considered as the cause of increasing trend of human CE caused by G6 in the country.
The cox1 sequences of G6 genotype in East Iran showed a higher similarity with isolates from Russia in North and Mongolia in Northeast Iran (Figure 1) (29), but more differences were found between isolates of East region and those from some other foci in Iran. Vast deserts such Kavir-e-Loot and Kavir-e-Namak separate East focus from other areas in Iran leading to a decrease in bio-relation between them. On the other hand, illegal influx of livestock including camels from Afghanistan, as the eastern neighbor of East region of Iran, has been reported for years. Although no study has been conducted on the genotyping of CE cases in Afghanistan, more political and social relationships of Afghanistan with Russia and East Asia may justify the similarity among G6 genotype of East Iran with those foci G6 isolates.
Based on the results of a study conducted in Argentina to investigate the association between clinical characteristics and Echinococcus genotypes, the early growth rate of G6 genotype in the liver of human cases was observed faster than other genotypes of Echinococcus, including G1 genotype (23). In addition, the rate of lung infection by genotype G6 in humans is more than that of other genotypes, as was the case in the present study. Moreover, in a study conducted by Sadjjadi and colleagues on human cerebral hydatid cysts in Iran, all investigated brain cysts (8/8) were identified as the G6 genotype of E. Canadensis, while the main cause of hydatid cyst in the study area was genotype G1 of E. granulosus (30). It can be concluded that G6 genotype of Echinococcus has more affinity to produce human cerebral hydatid cysts than other genotypes. The cerebral hydatid cysts are the most dangerous and deadly forms of hydatidosis. Since the cerebral hydatid cysts are slow growing and usually present late, the early diagnosis of these cysts is critical for treatment of the patients.
According to the dominance of G6 genotype of E. canadensis in the human cases of CE in Birjand, it is necessary to include appropriate methods for faster diagnosis and monitoring of the population at risk in the health care programs of this region.