1. Background
Different lactic acid bacterial strains including non - pathogenic Enterococci have a long history of safe use as microbial nutrition and probiotics (1). To a certain extent, Enterococci have been reported to produce low molecular weight proteins termed as bacteriocins. These bacteriocins exhibit antibacterial activity and are well - known to inhibit many food - borne pathogens and spoilage microorganisms. Hence, application of enterococcal bacteriocins enhances the safety of food products and extends their shelf life (2). Therefore, such enterococcal strains have widespread use in production of fermented foods and can be used safely for medical and veterinary applications (3). These Enterococci are considered as generally recognized as safe (GRAS) organisms as they are neither pathogenic nor opportunistic pathogens to man (4).
Also, Enterococci in yoghurt not only coagulate milk but also enhance desirable changes in taste, flavour, and texture of yoghurt (5). In such context, these bacteria have been studied by different yoghurt manufacturers in developed countries. However, little is known about the yoghurt Enterococci of Bangladesh although yoghurt is the most common fermented food product frequently consumed by the people of Bangladesh. Despite some studies showing the antibacterial activities of some yoghurt isolates (6), their potency or applicability against food - borne pathogens to prevent food - borne diseases has not been studied yet. In addition, these isolates were never been genotyped. In this study, Enterococci were isolated from yoghurt samples of Bangladesh and their antimicrobial properties were assessed in vitro against multidrug resistant bacteria isolated from food. Also, these isolates were genotyped for proper identification and attempts were made to preserve these strains, since these isolates could be one of the most important bio-properties of Bangladesh.
2. Methods
2.1. Collection of Yoghurt Samples and Isolation of Bacteria
Ten yoghurt samples were collected randomly from local markets and shops of Tangail, Bogra, Comilla, and Dhaka districts of Bangladesh. Bacteria were isolated from these yoghurts in MRS (de Man Rogosa Sharpe) medium (Oxoid, UK). Samples suspended in normal saline at 10% (w/v) were spread on MRS agar at pH 6.5 and were incubated anaerobically at 37 ºC overnight. Colonies were isolated from these plates and were transferred to MRS broth. Individual isolates were purified from the broth by streaking three times in MRS agar, and then were stored at -20 ºC and -80 ºC as glycerol stock.
2.2. Morphological and Biochemical Characterization of the Isolates
Gram staining was performed using standard procedure (7). The slides were observed under light microscope at 100X with oil immersion. Catalase test was performed by adding 3% hydrogen peroxide (Sigma, USA). Motility test was performed by stabbing bacterial cultures in semisolid MRS agar medium (0.5% agar) using a needle followed by incubation at 37 ºC overnight in anaerobic condition to observe diffusive zone of growth from the line of inoculation.
2.3. Molecular Characterization of the Isolates
For molecular characterization and species identification, part of 16S rDNA was amplified using the primers: Forward 5’ - TGGAGA GTTTGA TCCTGG CTCAG - 3’ and Reverse 5’ - TACCGC GGCTGC TGGCAC - 3’. For this, genomic DNA was isolated with phenol - chloroform - isoamyl alcohol method (6). The PCR condition includes an initial step of 5 min at 95 °C, then, 40 cycles of 30 seconds at 95 °C, 30 seconds at 55°C, and 1 min at 72 °C, followed by 10 min at 72 °C. After visualizing the PCR products on 1% agarose gel, the products were purified using PureLinkTM PCR purification kit and sequenced at the Genetic Engineering and Biotechnology Research Laboratory, Center for Advance Research in Science, University of Dhaka, Bangladesh. All the raw sequences were processed and aligned using Sequencher v5.4. These sequences were BLASTED against the 16S rRNA sequences database of the NCBI for species identification and were submitted to GenBank with referred accession numbers (KX646403 - KX646406). Nucleotide compositions of the processed sequences were analyzed using CLC Workbench v7.7.1 and Mega v5.05. Neighbour - joining (NJ) trees of K2P distances were created to provide a graphic representation of the patterning of divergence between species. Bootstrapping was performed in MEGA v5.05 with 1000 replications. All the statistical calculations were performed in Microsoft Excel.
2.4. Antibacterial Activity Assay
Antibacterial activity of the Enterococci was performed by agar well diffusion method as described before (8). Briefly, overnight culture of individual isolate in MRS broth was centrifuged at 10000 rpm for 20 minutes, and the cell free supernatant was used to perform antimicrobial activity against previously published multidrug resistant Shigella boydii, Klebsiella pneumoniae, Salmonella Typhimurium, E. coli 0157:H7, Staphylococcus aureus, Vibrio cholerae, and Enterobacter spp. isolated from food (9). Young cultures (McFarland standard 0.5) of these strains were spread on individual Muller Hilton agar (Oxoid, UK) plates, and wells were produced on plates using sterile yellow tips. Then, 20 µL of supernatant of each isolate was poured on each well and the plate was then incubated 4 ºC for 6 hours to allow the diffusion of the supernatant. Next, the plate was incubated at 37 ºC overnight to allow the bacterial growth, then, the diameter of the clear zone (≥ 5.0 mm) was measured with scale. The experiment was repeated three times.
2.5. Effect of Different Treatments on the Antibacterial Activity
Supernatants possessing antibacterial activity were treated with heat, proteinase K, or β - mercaptoethanol. For heat treatment, supernatant of cultured isolate was incubated at different temperatures (55, 70, and 90 ºC) for one hour in heat block and was then cooled at room temperature. For β - mercaptoethanol treatment, 78 µL of supernatant was added to 2 µL of β - mercaptoethanol (Sigma, USA). For proteinase K treatment, supernatant was treated with proteinase K (Sigma, USA) at a final concentration of 1 µg/mL overnight at 55 ºC, followed by inactivation of proteinase K at 95 ºC for 10 minutes and cooling at room temperature. Then, these treated supernatants were tested for antibacterial activity against different bacteria by agar well diffusion method as described above.
3. Results
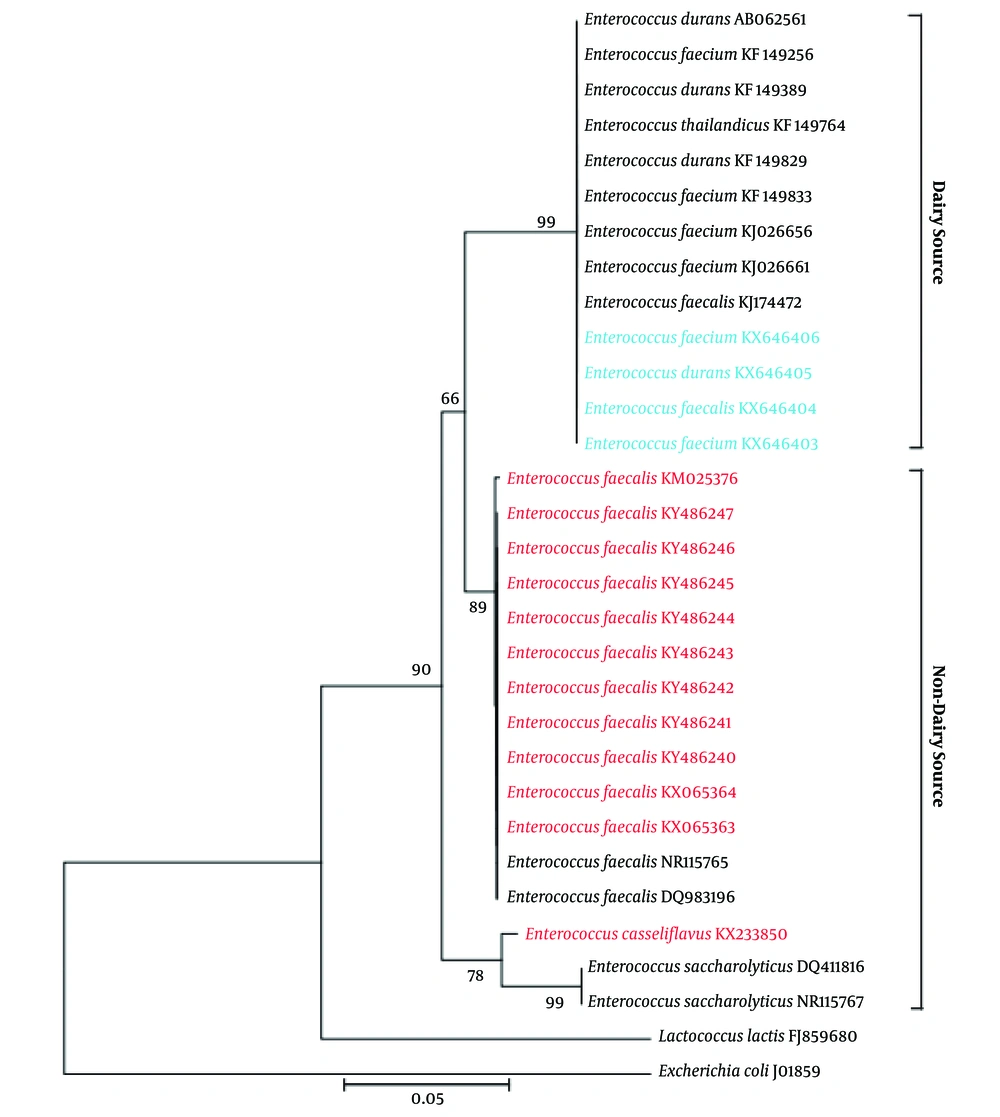
In this study, 18 enterococcal strains were isolated from 10 different yoghurt samples on MRS media based on their colony morphology (Table 1). All were Gram - positive, catalase negative, and nonmotile. Most of the cocci showed small, circular, and shiny colony, but cocci D02 was different, as it had irregularly shaped oily colony. Colony morphology of the diplococci CA0101 and TA0203 was mainly butyrous. 16S rDNA analysis revealed that all these strains were either Enterococcus faecalis, or Enterococcus faecium, or Enterococcus durans. The average nucleotide frequencies of the amplified 16S rDNA region was %A = 26.75 ± 0.85, %T = 20.17 ± 0.81, %G = 30.23 ± 1.16, and %C = 22.85 ± 0.44 with %AT = 46.6 ± 1.29 and %GC = 53.4 ± 1.29. Phylogenetic analysis revealed that the isolates clustered with other previously identified dairy samples but not with any non - dairy samples, either isolated from Bangladesh or abroad (Figure 1).
| Yoghurt Sample (Origin) | Isolates | Characteristics | Colony Morphology |
|---|---|---|---|
| A01 (Dhaka) | A0101 | +ve, Cocci | Small, circular, dry, shiny |
| A0102 | +ve, Streptococci | Very small, circular, dry, shiny | |
| A02 (Dhaka) | A0201 | +ve, Cocci | Small, circular, shiny |
| A0202 | +ve, Cocci | Small, circular, shiny, dry | |
| B (Bogra) | B0101 | +ve, Cocci | Small, opaque, shiny, pulvinate |
| B0102 | +ve, Cocci | Punctiform, convex with entire margin, opaque | |
| C01 (Comilla) | C0101 | +ve, Cocci | Medium sized, circular, dry, shiny |
| C03 (Comilla) | C0302 | +ve, Cocci | Punctiform, shiny |
| CA (Dhaka) | CA0101 | +ve, Diplococci | Medium sized, butyrous, rough surface |
| CA0102 | +ve, Cocci | Small, convex, opaque, dry, round, rough surface | |
| D01 (Dhaka) | D0101 | +ve, Cocci | Very small, convex, opaque |
| D0102 | +ve, Streptococci | Small, circular, shiny | |
| D02 (Dhaka) | D02 | +ve, Cocci | Irregular form, oily |
| TA01 (Tangail) | TA0101 | +ve, Cocci | Very small, shiny |
| TA0102 | +ve, Streptococci | Small, opaque, shiny | |
| TA02 (Tangail) | TA0201 | +ve, Cocci | Small, shiny |
| TA0202 | +ve, Cocci | Very small, shiny | |
| TA0203 | +ve, Diplococci | Punctiform, butyrous, circular, shiny |
When the cell - free cultured supernatant of these isolates were evaluated for antibacterial activities against food - borne pathogens by agar well diffusion method, variable spectrum of bacterial growth inhibition was observed. Supernatants from the Enterococci isolates C0302, CA0101, CA0102, and TA0203 showed the most antibacterial activity against all the seven pathogenic bacteria tested (Table 2). Among them, Enterococcus CA0101 showed the highest antibacterial activity, especially against V. cholerae with zone of inhibition (ZOI) 17.0 ± 0.0 mm, and its activity was the least for E. coli O157:H7, with a ZOI 7.5 ± 0.5 mm. Enterococcus C0302 also showed prominent antibacterial property against V. cholerae (ZOI 15.5 ± 0.5 mm). Enterococcus CA0102 showed highest antibacterial activity against V. cholerae and S. boydii (ZOI 10.5 ± 0.5 mm). Enterococcus TA0203 showed good activity against S. aureus and S. boydii (ZOI 12.5 ± 0.5 mm). Enterococci DO2 and TA0102 exhibited antibacterial activity against six and five out of the seven different pathogenic bacteria, respectively. The maximum ZOI found was 9.0 ± 0.5 mm against E. coli 0157:H7 by the Enterococci TA0102. A0202, B0102, TA0101, and TA0202 showed antibacterial activity against three different pathogens, whereas D0101, D0102, and TA0201 showed antibacterial activity against four different pathogens. However, the Enterococci isolates A0101, A0102, A0201, and C0101 did not show any antibacterial activities (Table 2). The degree of response of the pathogenic bacteria to the antibacterial activities of enterococcal isolates was in the order of E. coli 0157:H7 > V. cholerae > S. aurues > K. pneumoniae > S. Boydii > S. typhimurium > Enterobacter spp. (Table 2). This finding suggested that most of the isolates possessed antibacterial activity against food - borne pathogens.
| Isolates | Diameter of Zone of Inhibition (M ± SD) | ||||||
|---|---|---|---|---|---|---|---|
| Shigella boydii | Klebsiella pneumoniae | Salmonella typhimurium | E. coli O157:H7 | Staphylococcus aureus | Vibrio cholerae | Enterobacter spp. | |
| A0101 | - | - | - | - | - | - | - |
| A0102 | - | - | - | - | - | - | - |
| A0201 | - | - | - | - | - | - | - |
| A0202 | 6.5 ± 0.5 | 5.0 ± 0.5 | - | 5.0 ± 0.0 | - | - | - |
| B0101 | - | - | - | - | 9.5 ± 0.5 | 11.5 ± 0.5 | - |
| B0102 | - | - | - | 7.5 ± 0.5 | 8.0 ± 0.5 | 9.5 ± 0.5 | - |
| C0101 | - | - | - | - | - | - | - |
| C0302 | 9.5 ± 0.5 | 6.5 ± 0.5 | 9.5 ± 0.5 | 10.5 ± 0.5 | 11.5 ± 0.5 | 15.5 ± 0.5 | 6.0 ± 0.5 |
| CA0101 | 14.5 ± 0.5 | 9.0 ± 0.0 | 8.0 ± 0.0 | 7.5 ± 0.5 | 13.5 ± 0.5 | 17.0 ± 0.0 | 8.0 ± 0.5 |
| CA0102 | 10.5 ± 0.5 | 6.5 ± 0.5 | 7.0 ± 0.0 | 8.5 ± 0.5 | 9.0 ± 0.0 | 10.5 ± 0.5 | 5.0 ± 0.5 |
| D0101 | - | - | - | 7.5 ± 0.5 | 9.5 ± 0.5 | 15 ± 0.5 | 6.0 ± 0.5 |
| D0102 | - | - | 7.5 ± 0.5 | 7.0 ± 0.5 | 7.5 ± 0.5 | 10.5 ± 0.5 | - |
| D02 | 5.5 ± 0.5 | 5.0 ± 0.5 | 5.0 ± 0.5 | 8.5 ± 0.5 | 6.0 ± 0.5 | 8.0 ± 0.5 | - |
| TA0101 | - | - | 6.0 ± 0.0 | 6.5 ± 0.5 | - | 6.0 ± 0.5 | - |
| TA0102 | 8.5 ± 0.5 | 5.5 ± 0.5 | - | 9.0 ± 0.5 | - | 6.5 ± 0.5 | 5.0 ± 0.5 |
| TA0201 | 6.5 ± 0.5 | 5.5 ± 0.5 | - | 5.0 ± 0.5 | 7.0 ± 0.5 | - | - |
| TA0202 | 5.0 ± 0.5 | 5.0 ± 0.5 | - | 5.0 ± 0.0 | - | - | - |
| TA0203 | 12.5 ± 0.5 | 9.0 ± 0.0 | 9.5 ± 0.5 | 8.0 ± 0.5 | 12.5 ± 0.5 | 8.0 ± 0.0 | 5.0 ± 0.0 |
Abbreviation: SD, standard deviation.
The stability of the antibacterial activities of Enterococci isolates C0302, CA0101, CA0102, and TA0203 after different treatments was tested. After treating these supernatants at 55, 70, and 90 ºC, no significant change was observed in their antibacterial activity. Even after proteinase K and β - mercaptoethanol treatments, the antibacterial activity of the supernatant remained almost the same for each of the isolates (Table 3). These findings indicated that the supernatants might possess thermos - stable, proteinase K resistant, and disulphide bond - free antimicrobial compound or peptide (10).
| Isolates | Diameter of Zone of Inhibition (M ± SD) | ||||||
|---|---|---|---|---|---|---|---|
| Temperature (ºC) | PK + Supernatant | PK + Saline | BME + Supernatant | BME + Saline | |||
| 55 | 70 | 90 | |||||
| C0302 | 9.5 ± 0.5 | 9.5 ± 0.5 | 9.0 ± 0.5 | 9.0 ± 0.5 | 0.0 ± 0.0 | 9.0 ± 0.5 | 0.0 ± 0.0 |
| CA0101 | 14.0 ± 0.5 | 12.0 ± 0.5 | 10.0 ± 0.5 | 10.0 ± 0.0 | 0.0 ± 0.0 | 10.0 ± 0.5 | 0.0 ± 0.0 |
| CA0102 | 10.5 ± 0.5 | 10.5 ± 0.5 | 10.0 ± 0.5 | 10.0 ± 0.5 | 0.0 ± 0.0 | 10.0 ± 0.5 | 0.0 ± 0.0 |
| TA0203 | 12.5 ± 0.5 | 12.0 ± 0.5 | 12.0 ± 0.5 | 11.5 ± 0.5 | 0.0 ± 0.0 | 11.5 ± 0.5 | 0.0 ± 0.0 |
Abbreviations: SD, standard deviation; PK, proteinase K; BME, β - mercaptoethanol.
4. Discussion
Yoghurt is a common milk product consumed as a dessert in Bangladesh. Yoghurts rich in probiotics have wide acceptability because of their health benefits (4, 5). In this context, Lactobacilli and Lactococci were isolated from yoghurts of Bangladesh in some occasions (6), but there is no report on yoghurt Enterococci. In this study, we isolated and genotyped 18 Enterococci from 10 different yoghurt samples of Bangladesh. Morphology, growth characteristics, and general biochemical properties of these isolates were similar to previously isolated Enterococci from different sources (11). Moreover, analysis of partial 16S rDNA sequence revealed that they are either Enterococcus faecalis, or Enterococcus faecium, or Enterococcus durans, and none of them were Lactococcus. Thus, this study was the first to report yoghurt Enterococci from Bangladesh. Phylogenetic analysis showed that these isolates cluster with other Enterococci previously isolated from dairy sources, but not with the isolates from non - dairy sources (Figure 1). Such findings suggested that Enterococci from dairy and non-dairy sources are completely different strains and that dairy Enterococci might have some probiotic potentials.
Enterococci have the goodwill to produce bacteriocin, bacteriocin - like substances, or metabolites with antibacterial activity, which made them potential candidates for biopreservative agents (12, 13). Therefore, the cell free supernatants of these isolates were tested for antimicrobial activities, especially against food - borne pathogens. Ten of the isolates showed some degree of antibacterial activity only against Staphylococcus aereus, a Gram - positive bacterium. However, most of the isolates showed excellent antibacterial activity against all of the tested Gram - negative bacteria (Table 2). Such findings are comparable with previous reports (14-16). Interestingly, 13 Enterococci isolates showed potential antibacterial activity against E. coli 0157:H7, one of the frequent and highly pathogenic food - borne bacterium posing a potential threat to public health (17). Moreover, the antibacterial activity was not reduced when the supernatant was heated or treated with proteinase K, or even with β - mercaptoethanol. Possibly, the supernatant contained either antibacterial compound(s) or thermos- and proteinase K - resistant bacteriocin - like peptide(s) (10). Such findings offer promises to use these isolates as food preservatives. Further characterization and necessary modifications of these strains would be helpful to improve the quality of current yoghurt production.