1. Background
Staphylococcus aureus (S. aureus) is a Gram-positive cocci bacterium that colonizes different parts of the body such as nostrils and axilla (1). It colonizes about 25% - 35% of individuals and can cause infections in both communities and hospitals (1, 2). The microorganism can infect the skin, soft tissues, cardiovascular system, renal system, and respiratory system (1, 3, 4) and can cause toxic mediated infections such as toxic shock syndrome and sepsis (5, 6).
Methicillin-resistant S. aureus (MRSA) establishes resistance to beta-lactam antibiotics by expressing penicillin-binding protein-a2 (PBP2a) encoded by the mecA gene that alters the site for penicillin-binding on bacterial cell (7, 8). The gene is located on a staphylococcal cassette (SCCmec) sequence, which is of various subgroups (9). Then, S. aureus develops resistance to vancomycin, which is the drug of choice for the treatment of MRSA complicated infections (such a strain is named vancomycin-resistant S. aureus [VRSA]) (10).
There are a few scientific papers addressing the antibiotic profiles of S. aureus in Syria, which are frequently restricted to specific and limited laboratory centers in large cities (11). We found one study investigating the antimicrobial sensitivity pattern of S. aureus in Syria, and it was shown that 9.4% of the isolated strains were MRSA, and 1.9% were resistant to vancomycin (12). Therefore, data about antibiotic sensitivity patterns is sparse in this country.
2. Objectives
This study aimed to explore the prevalence rate of Staphylococcus aureus nasal carriage among secondary school students of Syrian civilians living in Duhok City, Iraq, and evaluate the susceptibility of S. aureus isolates to a range of antibiotics from different classes.
3. Methods
3.1. Study Setting
Ethical approval was obtained from different governmental entities and humanitarian agencies including the Ethics Committee of the College of Medicine (no.: 22112017-7; 22nd /Nov./2017), The University of Duhok, Board of Relief and Humanitarian Affairs for refugees and internally displaced people in Duhok (no.: 2926; 23/Nov./2017), and United Nations High Commissioner for Refugees (code: HCR/IRQ/DHK/2017/L/010; 1st/Feb./2017).
As Duhok city hosts 109,352 registered Syrian refugees (13), this cross-sectional and community-based study was carried out in Duhok in the Kurdistan Region in Northern Iraq. We collected samples from four camps of Syrian refugees and a group of immigrants upon arrival at the border entry point to Iraq. The present study was conducted between January and August 2019.
3.2. Exclusion Criteria and Sample Collection
Samples were collected from Syrian students attending schools inside the camps. Before sample collection, signed consents were obtained from students/guardians. Subjects with one of the following criteria were excluded: recent infection, previous admission to hospital, admission for surgical procedures, renal dialysis, use of a catheter or intravenous line at the time of swab taking, former detection of MRSA from the subject, and skin infections (6).
Samples were collected from the nostrils of the participants by using sterile swabs after moistening them with sterile normal saline. Swabs were inserted into the nares for about 2 cm and rolled gently for a few seconds. Then, swabs were inoculated into brain heart infusion (BHI) media (Neogen, UK) and transported to the lab for microbiological investigations.
3.3. Laboratory Practice
3.3.1. Isolation and Identification of S. aureus
Several laboratory tests were performed for the isolation and identification of S. aureus isolates. After the samples were transported to the lab using BHI and incubated for about 16 - 18 h aerobically at 37°C, they were cultured on selective medium (mannitol salt agar; Neogen company, UK) for the isolation of S. aureus and incubated aerobically overnight at 35°C - 37°C. Subsequently, plates were observed for colony phenotypic characteristics, followed by Gram staining for studying cellular characteristics of the microorganism according to Barrow et al. (14). Further, the isolates were differentiated from other Gram-positive cocci bacteria by performing catalase and tube coagulase tests (14).
3.3.2. Antibiotic Susceptibility Testing for S. aureus Isolates
Kirby-Bauer disk diffusion method and agar dilution assay, for oxacillin and vancomycin, was used for conducting the antibiotic sensitivity tests. Various antibiotics from different classes were tested such as oxacillin (6 µg/mL), vancomycin (30 µg/mL), tetracycline (30 µg), teicoplanin (30 µg), gentamicin (10 µg), clindamycin (2 µg), rifampicin (5 µg), ciprofloxacin (5 µg), and fusidic acid (10 µg). In addition, Muller Hinton Agar (MHA) (Neogen, UK) was used in conducting the test. The antibiotic sensitivity test for the mentioned antibiotics was conducted according to the Clinical and Laboratory Standards Institute as well as the results interpretation (15). The procedure for the Kirby-Bauer disk diffusion method included inoculation of bacterial suspension with a McFarland of 0.5. Then, bacteria were cultured on MHA using a sterile swab, and antibiotic disks were applied to the plate and incubated overnight (18 - 24 h) at 35°C. The agar dilution assay, for oxacillin and vancomycin, was performed by adding the antibiotics into the agar itself and plated the bacteria on the plate and incubated overnight at 35°C.
3.4. Statistical Analysis
Data is presented as frequencies and percentages. Minitab software version 17 was used for data analysis.
4. Results
The total number of interviewed participants was 426; among them, 71 of the participants were excluded because they met one of the aforementioned exclusion criteria. Consequently, 355 samples were collected and analyzed. The participants were aged 16 years or older, and 114/335 (34%) were male.
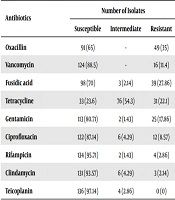
Out of the 355 samples, 140 (39.4%) were confirmed as S. aureus carriers, and the highest resistance rate was against oxacillin 49/140 (35%) followed by fusidic acid (27.86%). The resistance rates against tetracycline, gentamicin, vancomycin, and ciprofloxacin were 22.1%, 17.86%, 11.4%, and 8.57%, respectively. All the strains were sensitive to teicoplanin (Table 1).
| Antibiotics | Number of Isolates | ||
|---|---|---|---|
| Susceptible | Intermediate | Resistant | |
| Oxacillin | 91 (65) | - | 49 (35) |
| Vancomycin | 124 (88.5) | - | 16 (11.4) |
| Fusidic acid | 98 (70) | 3 (2.14) | 39 (27.86) |
| Tetracycline | 33 (23.6) | 76 (54.3) | 31 (22.1) |
| Gentamicin | 113 (80.71) | 2 (1.43) | 25 (17.86) |
| Ciprofloxacin | 122 (87.14) | 6 (4.29) | 12 (8.57) |
| Rifampicin | 134 (95.71) | 2 (1.43) | 4 (2.86) |
| Clindamycin | 131 (93.57) | 6 (4.29) | 3 (2.14) |
| Teicoplanin | 136 (97.14) | 4 (2.86) | 0 (0) |
aValues are expressed as No. (%).
5. Discussion
In this study, the prevalence rate of nasal carriage of S. aureus was similar to the nasal carriage rate in Italy (39.2%) (16). This rate was lower than what was found in previous studies in Syria (48%) (12) and Ethiopia (47.74%) (17). However, our results were higher than what was found in Iraq (17.5% - 30%) (6, 18-20), Iran (28%) (21), India (25%) (22), Thailand (29.7%) (23), and Malaysia (31%) (24). This variation in rates among different populations might be due to the differences in standards used in performing the lab work and sample size.
We found that the highest rate of antibiotics resistance was against oxacillin (35%), and this was higher than what was found in previous studies conducted in Syria (9.4%) (12) and Iraq (2.04% - 21.95%) (6, 19, 20), and no oxacillin resistant isolates were found in Malaysia (24). The rate was lower than that in Nigeria (47.15%) (25), which might be due to the misuse of antibiotics especially in developing countries where individuals can obtain antibiotics easily and without prescription.
Vancomycin serves as an alternative drug of choice for treating MRSA infections. The resistance rate of S. aureus against vancomycin in the present study was 11.4%. This rate was higher than what was found in Syria (1.9%) (12) and China (4.4%) (26). Besides, it was also observed that no resistance was found against vancomycin in different studies in Iraq (5, 20, 27), Ethiopia (17), and the United States of America (USA) (28). With the high rate of resistance to vancomycin, it increases the challenge of treating infections caused by multi-drug resistant S. aureus.
On the other hand, we found a lower resistance rate to tetracycline than what was found in Iraq (25%) (27) and Thailand (34.2%) (23), while it was higher than the reports from Nepal (20%) (5), Malaysia (17.3%) (24), and Ethiopia (16.9%) (17).
Furthermore, we found that the resistance rate to gentamicin was 17.86%, which is higher than those rates reported in Iraq (3.22%) (27) and Ethiopia (1.5 %) (17) and lower than the reports from Nepal (33.3%) (5) and USA (30%) (28). Furthermore, about 8.57% of S. aureus isolates showed resistance to ciprofloxacin. This rate was lower than what was found in Nepal (36.7%) (5), Iraq (19%) (27), and the USA (65%) (28). However, another study conducted in Iraq showed that all isolates were sensitive to ciprofloxacin (20).
The resistance rates of S. aureus to rifampicin and clindamycin were 2.86% and 2.14%, respectively. The rifampicin resistance rate in the present study was higher than what was found in the USA (0%) (28). However, the clindamycin resistance rate (2.14%) was lower than the results of a study conducted in Syria (21.64%) (12), Nepal (13.3%) (5), Thailand (63.2%) (23) and USA (57%), while the resistance rate was similar to the rate reported in Malaysia (2.2%) (24). In addition, we found that all S. aureus isolates were sensitive to teicoplanin. Our finding was similar to what was found in Nepal (5). This similarity is difficult to explain, and further research is needed to explore this. The variation in resistance rates to different types of antibiotics included in the study could be due to the variation in different factors such as the age group of participants, gender, and laboratory standards. Besides, the misuse of antibiotics plays an important role in spreading the resistance genes among the population.
However, our study recruited a limited number of refugees on the point of arrival in Iraq. This was due to the difficulty in accessing Iraqi-Syrian borders due to regional insecurity. Thus, further studies recruiting larger sample sizes are recommended to determine the concise status of the prevalence rate of microorganisms in the Syrian community.
5.1. Conclusions
This study focused on finding the nasal carriage rate and determining the antibiotics susceptibility profile of S. aureus isolates among Syrian refugees in Iraq. The highest rate was against oxacillin, and all isolates were sensitive to teicoplanin. Vancomycin susceptibility testing showed a significant rate of resistance, and this increased the challenge of treating infections caused by multi-drug resistant S. aureus. Further studies are needed to investigate the molecular base of antibiotics resistance in this microorganism.