1. Background
Urinary tract infection (UTI) is one of the most common bacterial diseases (1). Globally, 150 million people are diagnosed with UTI annually (2), which costs more than 6 billion US dollars. UTIs are a spectrum of diseases ranging from simple cystitis to serious infections such as pyelonephritis and other complications in humans (3). In general, UTI is more common in females than in males, as the female urethra is structurally found less effective to prevent the bacterial entry (4). Besides, the frequency rate of UTI depends on different factors, including age, previous use of antibiotics, hospitalization, and catheterization.
It is known that more than 95% of urinary tract infections are caused by a single bacterial species. E. coli is the most frequent infecting organism in acute infections (5). In recent reports carried out in Iraq, it was found that E. coli, Staphylococcus spp., and K. pneumonia were the most common infectious agents causing UTI, which were also resistant to the most commonly used antibiotics (6, 7).
The early treatment of UTI with empirical antibiotics decreases the rate of morbidity (8). In order to administer an appropriate empirical therapy, it is critical to know the main bacteria causing urinary tract infection as well as their respective antimicrobial susceptibility pattern (9). Determining microorganisms and their antibiotic sensitivity patterns allows good treatment outcomes, controls the increase of antimicrobial prescription and helps to control antimicrobial resistance, which is a public health problem worldwide. In addition, pathogens recovered from males were shown to be more resistant to antibiotics when compared with pathogens isolates from female patients. Thus, while choosing an empirical antimicrobial therapy, gender should be taken into account (10).
2. Objectives
In this study, we aimed to evaluate the distribution of pathogens associated with UTI in male patients and determine the pathogens’ antimicrobial susceptibility patterns in Duhok Province, Iraq.
3. Methods
The present study was approved by the Ethics Committee of the College of Medicine, University of Zakho, Kurdistan, Iraq (code: 4/154/NW/02.05.2019) and was carried out in Duhok Province, Kurdistan Region, Iraq from January 2017 to February 2020. All patients (aged 10 - 65 years old) completed the consent form before being recruited in the study. Inclusion criteria included male gender, positive microbiological evidence of UTI (bacterial growth of higher than 105 CFU/mL), and willingness to be recruited in the study.
A total of 211 urine samples were collected from male patients with UTI visiting private health centers. Midstream-clean catch urine was collected from patients in a sterile disposable container (5 mL) to avoid contamination. Then, the urine samples were inoculated on Blood and MacConkey agars (Oxoid Ltd, Bashingstore, Hampire, UK) and incubated at 37°C for 24 h. Cultures showed no growth at the end of 24 h incubation and were further incubated for 48 h. Bacterial isolates were initially classified using Gram-staining, and then the isolates were identified depending on the standard microbiological culture and biochemical characteristics. The identification of bacteria was performed according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (11, 12). Vitek-2 Microbial Analysing system (bioMerieux, US) was used for antimicrobial sensitivity testing.
4. Results
4.1. Pathogens
A total of 211 male subjects met our inclusion criteria and were recruited in the study. Out of these, 170 (80.6%) isolates were Gram-negative bacteria, and 41 (19.4%) were Gram-positive (Table 1). The most commonly isolated organisms were E. coli (52.6%), P. aeruginosa (14.2%), Klebsiella pneumonia (6.2%), Staphylococcus haemolyticus (5.7%), and Staphylococcus epidermidis (4.3%) (Table 1).
| Isolated Bacteria | Valuea |
|---|---|
| Gram negative bacteria (n = 170 [80.6%]) | |
| Escherichia coli | 111 (52.6) |
| Pseudomonas aeruginosa | 30 (14.2) |
| Klebsiella pneumonia | 13 (6.2) |
| Proteus mirabilis | 9 (4.3) |
| Morganella morganii | 4 (1.9) |
| Actinobacter baumanni | 3 (1.4) |
| Gram positive bacteria (n = 41 [19.4%]) | |
| Staphylococcus haemolyticus | 12 (5.7) |
| Staphylococcus epidermidis | 9 (4.3) |
| Enterococcus faecalis | 6 (2.8) |
| Streptococcus agalactiae | 6 (2.8) |
| Staphylococcus lentus | 5 (2.4) |
| Staphylococcus aureus | 3 (1.4) |
| Total | 211 (100) |
Frequency of Bacterial Isolates From Urine Specimens in this Study (Male = 211)
4.2. Bacterial Susceptibility
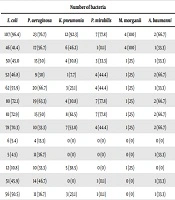
The comparison of the susceptibility pattern of Gram-negative and -positive pathogens to various antimicrobial agents from all the specimens is summarized in Tables 2 and 3, respectively. E. coli showed high resistance to ampicillin (96.4%), cefepime (72.9%), ceftriaxone (72.1%), and aztreonam (70.3%). There was a high susceptibility pattern of E. coli to imipenem (95.5%) and ertapenem (94.6%). P. aeruginosa showed high resistance to ciprofloxacin (83.3%), and ampicillin (76.7%), while it was highly sensitive to ertapenem (86.7%). Additionally, K. pneumonia was highly susceptible to imipenem (100%), gentamicin (100%), and ertapenem (100%), and 92.3% of the isolated strains were resistant to ampicillin. It was also found that around 78% of Proteus mirabilis isolates were resistant to ampicillin, cefepime, and ceftriaxone, and 100% were susceptible to imipenem, amikacin, and ertapenem (Table 2).
| Antibiotic | Number of Bacteria | |||||
|---|---|---|---|---|---|---|
| E. coli | P. aeruginosa | K. pneumonia | P. mirabilis | M. morganii | A. baumanni | |
| Ampicillin | 107 (96.4) | 23 (76.7) | 12 (92.3) | 7 (77.8) | 4 (100) | 2 (66.7) |
| Amoxicillin/clavulanic acid | 46 (41.4) | 17 (56.7) | 6 (46.2) | 1 (11.1) | 4 (100) | 1 (33.3) |
| Piperacillin/tazobactam | 50 (45.0 | 15 (50) | 4 (30.8) | 3 (33.3) | 1 (25) | 1 (33.3) |
| Cefazolin | 52 (46.8) | 9 (30) | 1 (7.7) | 4 (44.4) | 1 (25) | 2 (66.7) |
| Cefixime | 62 (55.9) | 20 (66.7) | 3 (23.1) | 4 (44.4) | 1 (25) | 1 (33.3) |
| Ceftriaxone | 80 (72.1) | 19 (63.3) | 4 (30.8) | 7 (77.8) | 1 (25) | 2 (66.7) |
| Cefepime | 81 (72.9) | 15 (50) | 8 (61.5) | 7 (77.8) | 1 (25) | 2 (66.7) |
| Aztreonam | 78 (70.3) | 10 (33.3) | 7 (53.8) | 4 (44.4) | 1 (25) | 2 (66.7) |
| Ertapenem | 6 (5.4) | 4 (13.3) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Imipenem | 5 (4.5) | 11 (36.7) | 0 (0) | 0 (0) | 0 (0) | 1 (33.3) |
| Amikacin | 12 (10.8) | 10 (33.3) | 5 (38.5) | 0 (0) | 1 (25) | 0 (0) |
| Gentamicin | 51 (45.9) | 14 (46.7) | 0 (0) | 1 (11.1) | 0 (0) | 1 (33.3) |
| Tobramycin | 56 (50.5) | 11 (36.7) | 3 (23.1) | 1 (11.1) | 0 (0) | 1 (33.3) |
| Ciprofloxacin | 70 (63.1) | 25 (83.3) | 5 (38.5) | 2 (22.2) | 0 (0) | 2 (66.7) |
| Levofloxacin | 66 (59.5) | 21 (70) | 4 (30.8) | 4 (44.4) | 0 (0) | 1 (33.3) |
| Nitrofurantoin | 18 (16.2) | 8 (26.7) | 6 (46.2) | 4 (44.4) | 0 (0) | 1 (33.3) |
| Trimethoprim/sulfamethoxazole | 61 (54.9) | 7 (23.3) | 9 (69.2) | 4 (44.4) | 0 (0) | 1 (33.3) |
| Antibiotic | Number of Bacteria | |||||
|---|---|---|---|---|---|---|
| S. haemolyticus | S. epidermidis | E. faecalis | S. agalactiae | S. aureus | S. lentus | |
| Benzylpenicillin | 12 (100) | 9 (100) | 5 (83.3) | 4 (80) | 2 (66.7) | 5 (100) |
| Oxacillin | 11 (91.7) | 7 (77.8) | 5 (83.3) | 3 (60) | 0 (0) | 5 (100) |
| Gentamicin | 2 (16.7) | 1 (11.1) | 2 (33.3) | 1 (20) | 0 (0) | 2 (40) |
| Tobramycin | 6 (50) | 3 (33.3) | 1 (16.7) | 3 (60) | 0 (0) | 2 (40) |
| levofloxacin | 8 (66.7) | 1 (11.1) | 5 (83.3) | 2 (40) | 0 (0) | 3 (60) |
| Moxifloxacin | 3 (25) | 1 (11.1) | 5 (83.3) | 2 (40) | 1 (33.3) | 2 (40) |
| Erythromycin | 10 (83.3) | 4 (44.4) | 5 (83.3) | 2 (40) | 0 (0) | 3(60) |
| Clindamycin | 6 (50) | 3 (33.3) | 5 (83.3) | 5 (100) | 0 (0) | 5 (100) |
| Linezolid | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (60) |
| Teicoplanin | 2 (16.7) | 2 (22.2) | 2 (33.3) | 0 (0) | 0 (0) | 1 (20) |
| Vancomycin | 2 (16.7) | 1 (11.1) | 0 (0) | 0 (0) | 0 (0) | 2 (40) |
| Tetracycline | 6 (50) | 4 (44.4) | 5 (83.3) | 0 (0) | 0 (0) | 2 (40) |
| Tigecycline | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Nitrofurantoin | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Rifampicin | 1 (8.3) | 1 (11.1) | 0 (0) | 3 (60) | 0 (0) | 1 (20) |
| Trimethoprim/sulfamethoxazole | 5 (41.7) | 2 (22.2) | 3 (50) | 1 (20) | 1 (33.3) | 0 (0) |
Staphylococcus spp. were responsible for about 13.8% of UTI cases. Out of these, S. haemolyticus (5.7%) and S. epidermidis (4.3%) were the most frequent Gram-positive bacteria, which showed high resistance to benzylpenicillin (100%) and 100% susceptibility to nitrofurantoin, linezolid, and tigecycline (Table 3). In addition, around 83% of the isolated E. faecalis strains were resistant to benzylpenicillin, oxacillin, levofloxacin, moxifloxacin, erythromycin, and clindamycin, but highly sensitive to linezolid (100%), nitrofurantoin (100), tigecycline (100%), rifampicin (100%), and vancomycin (100%) (Table 3). The resistance rates of S. agalactiae, S. lentus, and S. aureus to other commonly used antibiotic agents are presented in Table 3.
5. Discussion
UTI is one of the most widespread infections worldwide (1). E. coli is considered as the most frequent uropathogen involved in community-acquired UTI (being implicated in more than half of all the UTI cases) (13). The prevalence of UTI varies according to gender, age, geographical and regional locations, previous use of antibiotics, hospitalization, and catheterization (9).
Geographic variations in pathogen occurrence and antibiotic susceptibility profiles require frequent monitoring to provide information to guide the therapeutic options. Therefore, this study aimed to investigate the frequency of microorganisms responsible for UTIs and their antimicrobial susceptibility patterns in males in Duhok Province, Iraq. The majority of studies have reported that Gram-negative bacteria cause 90% of UTI cases, while Gram-positive bacteria cause only 10% of the cases (14, 15). In the present study, we found that Gram-negative bacteria were a common cause of UTIs among males (80.6%); this was in agreement with reports of other studies conducted in Iran and Iraq (3, 6, 16). This could be due to the presence of unique structure in Gram-negative bacteria, which facilitates attachment to the uroepithelial cell, resulting in high prevalence in UTIs.
Following the classification of the detected pathogenic bacteria, our data showed that among the isolated Gram-negative pathogens, E. coli was the most frequent isolate, and A. baumanni was the least detected one in our community. E. coli is the most common isolated etiological agent responsible for 80% - 90% of uncomplicated UTIs, which could be due to the fact that it belongs to the normal flora of the human intestine, and therefore, it easily colonizes the urinary tract and can exhibit multidrug resistance (15). Likewise, other studies reported that E. coli was the most common pathogen causing UTIs in males (17, 18). Likewise, several previous studies conducted in Iraq showed that E. coli was the most common causative microorganism responsible for UTIs (7, 19).
However, the resistance pattern of E. coli to antibiotics has been very different in various studies. In a study conducted in Iran 2006, Sharifian et al. found the highest susceptibility rate of E. coli to ceftriaxone (97.8%) and cefotaxime (95.2%) (20). Other studies carried out in Iraq reported that E. coli was highly sensitive to imipenem and meropenem (16). In this study, however, E. coli showed high sensitivity to imipenem and ertapenem. This could be due to the antimicrobial agents used in the community under medical prescription. However, E. coli showed full resistance to ampicillin, cefepime, ceftriaxone, and aztreonam. Such high resistance rates to antibiotics in our community can be explained partially by the high rate of antibiotics abuse in the region.
Our results also showed that P. aeruginosa was the second the most common bacterium that caused UTI in males, which is in agreement with the findings of a previous study in Iran (11). Another report from Ethiopia found that P. aeruginosa was the second most common cause of UTI in males (21). The bacterium P. aeruginosa is emerging as an opportunistic pathogen of UTI in the community and has been associated with 10.0% - 25% of male cases (8). The prevalence rate P. aeruginosa in the present study was higher than that reported in other studies conducted in Turkey (22), Iran (23), and Italy (4). Such variations reported in different studies might be due to the differences in sample collection, study design, and inclusion criteria. The sensitivity pattern of P. aeruginosa was alarming as the vast majority of isolated strains in this study were resistant to all the commonly used antibiotics, which is in line with reports from other studies conducted in Iraq (6, 16).
In our study, K. pneumonia was shown to be responsible for about 6.2% of UTI cases and was highly sensitive to imipenem (100%), gentamicin (100%), and ertapenem (100%), while 92.3% of the isolated strains were resistant to ampicillin. This is in agreement with previous findings indicating K. pneumonia as a common cause of UTI (24, 25). Additionally, we found that more than two-thirds of P. mirabilis isolates were resistant to ampicillin, cefepime, and ceftriaxone, while 100% of the isolates were susceptible to imipenem, amikacin, and ertapenem. Furthermore, M. morganella and A. baumanni were the least frequent causes of UTI among males in the present study. These organisms showed high resistance to commonly used antibiotics such as ampicillin (100%) and amoxicillin/clavulanic acid (100%) and high susceptibility to most of the antibiotics tested, which is in agreement with data published by others (19).
In terms of Gram-positive bacteria, Staphylococcus spp. were responsible for about 13.8% of UTI cases, including S. haemolyticus (5.7%), S. epidermidis (4.3%), and S. aureus (1.4%). In agreement with this finding, previous studies reported that Staphylococcus spp. as the most common causative agents among Gram-positive bacteria causing UTIs (7, 19). In a previous study in Iraq, it was found that about 50% of the bacteria causing UTIs were Gram-positive, and the majority of them were Staphylococcus spp. (6). Both S. haemolyticus and S. epidermidis showed high resistance rates to benzylpenicillin (100%). These strains were 100% susceptible to nitrofurantoin, linezolid, and tigecycline. Additionally, S. aureus was highly sensitive to all the commonly used antibiotics in this study. In contrast, other studies conducted in Iraq showed that S. aureus was resistant to the most commonly used antibiotics (26-28).
In this study, a few cases of UTI resulted from S. agalactia showing 100% susceptibility to linezolid, teicoplanin, vancomycin, tetracycline, tigecycline, and nitrofurantoin and high resistance to benzylpenicillin (80%). This was in agreement with a previous study that demonstrated that S. agalactiae was less frequently associated with UTI, with an isolation rate ranging from 3.96% to 5.7% (8). Furthermore, these results were similar to other studies that found that S. agalactiae strains were highly sensitive to vancomycin and nitrofurantoin (29, 30). In the present study, E. faecalis was responsible for 2.8% of the UTI cases. Antibiotic susceptibility test showed that E. faecalis was highly resistant to benzylpenicillin, oxacillin, levofloxacin, moxifloxacin, erythromycin, clindamycin, and tetracycline. These findings were in contrast to the results of a previous study conducted in Iraq, recruiting 151 subjects of both genders, which reported that E. faecalis was the second most common infectious agent causing UTIs (23.4%) (6). These differences could be attributed to the differences in sample size, study design, inclusion, and exclusion criteria.
The small number of samples was one of the limitations of the present study that may not show the actual amount of occurrence in the population. Secondly, the study was conducted mainly among male patients, and age groups less than 10 years were not analyzed in the study. Therefore, further studies using a molecular technique to diagnose and evaluate the sensitivity of bacteria should be conducted in the region to overcome these limitations.
5.1. Conclusions
From the results of the present study, it is concluded that the main pathogen causing UTIs among males is E. coli, followed by P. aeruginosa and Staphylococcus spp. Our results showed that the majority of isolates were resistant to commonly prescribed antibiotics such as ampicillin, ceftriaxone, cefepime, benzylpenicillin, oxacillin, and erythromycin. This is an alarming situation, and an urgent plan to control this threatening development of antibacterial resistance is required.