1. Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first reported to the World Health Organization from Wuhan, China, on December 31, 2019 (1, 2). Two coronavirus epidemics have occurred in the last 20 years. The first one is severe acute respiratory syndrome (SARS) coronavirus which started in China affecting 10 countries with about 8000 cases and 800 mortalities. The second one is Middle East respiratory syndrome coronavirus emerging from Saudi Arabia with about 2500 cases and 800 mortalities (3, 4). In Iran, on February 19, 2020, the first two cases of coronavirus disease 2019 (COVID-19) were reported (5).
Coronavirus is a ribonucleic acid (RNA) virus that is recognized by the presence of glycoproteins on its surface (corona meaning crown in Latin). The transmission of this virus is similar to that of the SARS virus but more probable than the flu (6). The incubation period of the disease is 6 days on average and can last up to 14 days. Patients with minimal symptoms can also transmit the disease; however, the extent of this transfer is not yet well known (7). The clinical manifestations of COVID-19 range from asymptomatic to respiratory failure requiring mechanical ventilation and intensive care (8).
The first report of clinical signs of the disease, by Huang et al. in Wuhan, including fever, dry cough, weakness, and shortness of breath, were the most common signs and symptoms. Moreover, in all cases, chest computed tomography (CT) scans showed parenchymal involvement. About 32% (n = 13) of these patients needed to be admitted to the intensive care unit, and 15% (n = 6) died (9). These symptoms were more common in patients with diabetes and hypertension than in other patients (10). However, in a study of 24 patients with asymptomatic infections, 50% of patients had typical lung consolidation in the chest CT, and another 20% had atypical imaging abnormalities (11).
In SARS-CoV-2 patients, the number of white blood cells (WBCs) can vary. Leukopenia, leukocytosis, and lymphopenia have been reported; however, lymphopenia is the most common. High levels of lactate dehydrogenase and ferritin are common, and high aminotransferase levels have been described. At the time of admission, numerous patients with pneumonia have normal serum procalcitonin levels. High levels of D-dimer and more prominent lymphopenia have been associated with higher mortality (12-15). During SARS-CoV-2 infection, the frequency of naïve B cells has been reported to be reduced; nevertheless, the plasma cells have been observed to remarkably increase in peripheral blood mononuclear cells. Additionally, given the pivotal role of B cells in the control of infections, the positive detection of immunoglobulin M (IgM) and immunoglobulin G (IgG) antibodies specific to SARS-CoV-2 is an important process for the clinical evaluation of infections (16).
For COVID-19 diagnosis, reverse transcription polymerase chain reaction (RT-PCR) is used as a reference test, through which SARS-CoV-2 is detected (17). However, in individuals with suspected imaging findings, it might yield a false negative result (18), which could be due to the incorrect sampling of the upper respiratory tract (nasopharynx and pharynx) or incorrect extraction technique of nucleic acid (19). The final diagnosis of this disease is made by RT-PCR (20). If the initial test is negative, there is still suspicion that resampling should be performed from different parts of the respiratory tract.
Patients after a partial recovery are discharged and recommended to be quarantined at home (21). These patients might transfer the infection when they become asymptomatic. These partially recovered patients might be a great source for the virus spread. The pandemic has gone much further than what was expected and has become the greatest challenge of the century.
2. Objectives
The current study aimed to understand when RT-PCR results would be negative after recovery of signs and symptoms in SARS-CoV-2 patients that help prevent the disease spread by partially recovered patients to some extent.
3. Methods
This cross-sectional study was performed on a total of 10 patients with mild to moderate illness referring to Kowsar Hospital in Semnan, Iran, who met the inclusion criteria after receiving informed written consent. The patients underwent a spiral chest CT scan if they had clinical signs and symptoms. If the CT had positive findings suggestive of COVID-19 infection, serum acute phase C-reactive protein (CRP) and then an RT-PCR test were performed on nasopharyngeal and oropharyngeal samples. If the RT-PCR result was positive, the patient entered the investigation. The inclusion criteria were informed written consent, CT scan findings in favor of COVID-19 (bilateral pulmonary parenchymal ground-glass and pulmonary consolidations and opacities) (22), presence of clinical symptoms up to 48 hours before RT-PCR, CRP positive result, age range of 25 - 65 years, no underlying disease (eg, diabetes, hypertension, and heart disease), and positive results in RT-PCR. The exclusion criteria were patients’ traveling, not being in quarantine at home, using other medications recommended by an infectious disease specialist, worsening clinical signs and symptoms needing hospitalization, and not giving consent to follow-up and repeated RT-PCR or incomplete follow-up checklists.
The demographic characteristics of the patients, including age, gender, signs and symptoms, CRP, and WBC findings, were recorded. The patients were asked to fill in the checklists to record their daily signs and symptoms and their auxiliary body temperature by a digital thermometer. The patients were followed daily through a phone call to ensure from filling the checklists appropriately, becoming aware of their general condition, and answering their questions. The RT-PCR test was performed every 3 days for all patients. After 2 days of cessation of clinical symptoms, a nasopharyngeal and oropharyngeal sample was taken. If the patients’ test had a negative result, a repeated RT-PCR was performed 24 hours later. If two consecutive tests had negative results, the patient was considered definitively treated (23), and the follow-up was terminated. Moreover, if the RT-PCR test was positive, the test was performed 3 days later. All patients received the same therapeutic regimen by an infectious disease specialist, including hydroxychloroquine 200 mg every 12 hours up to 5 days, naproxen 250 mg every 12 hours up to 5 days, azithromycin 500 mg for the first day, and then 250 mg daily for 5 days, and pantoprazole 40 mg daily.
The patients were able to contact the researcher at any time. Real-time PCR was performed on the samples obtained from nasopharyngeal and oropharyngeal swabs collected in sterile viral transport medium tubes by a physician and nurse. Viral RNA was extracted in an automated manner, and complementary deoxyribonucleic acid was obtained. Automatic RNA extraction was performed using magnetic-particle technology (MagCore®, Taiwan) and real-time PCR using the StepOnePlus (Applied Biosystems, USA). Briefly, N- and ORF1ab-gene of SARS-CoV-2 was quantified by real-time RT-PCR on both nasopharyngeal and oropharyngeal swabs using a Detection kit for the 2019 novel coronavirus RNA (PCR-Fluorescence Probing) (Daan Gene Co., Ltd. Sun Yat-Sen University, Guangdong, China) according to the manufacturer’s protocol. Both N- and ORF1ab- genes of each individual sample were measured in the FAM (Fluorescein amidites) and VIC (Victoria) detecting channels, respectively. Cycle threshold values ≤ 40 were considered positive.
For the detection of SARS-CoV-2 IgM and IgG antibodies, blood specimens were obtained from patients approximately 1 month after symptom relief. An indirect enzyme-linked immunosorbent assay was used to qualitatively detect IgM and IgG antibodies against the N (nucleocapsid) antigen of SARS-CoV-2, according to the manufacturer’s detailed instruction (Pishtaz Teb Diagnostics Co., Iran). Briefly, specific antibodies in serum collected from participants react with solid-phase antigens. Subsequently, the secondary conjugated antibody containing antihuman IgM/IgG horseradish peroxidase conjugate was added to the wells. Following the addition of chromogen-substrate, the development of yellow color was measured spectrophotometrically at 450 nm. The qualitative results were expressed as a cut-off index (COI), with a COI < 0.9 and > 1.1 indicating negative and positive results, respectively. Considering clinical findings, lung RT-CT positive results, and a positive RT-PCR test, the manufacturer reported sensitivity of 79.4% and 94.1% and specificity of 97.3% and 98.3% for IgM and IgG, respectively.
The data were analyzed using SPSS Software (version 20, SPSS Inc., Chicago, USA). Pearson correlation was used to assess the relationship between each of the independent variables with the time to reach a negative RT-PCR result. A P-value less than 0.05 was considered statistically significant. The study was approved by the Ethics committee of Semnan University of Medical Sciences (IR.SEMUMS.REC.1399.011). The patients were ensured regarding the confidentiality and anonymity of their information. The patients were free to withdraw from the study at any step without affecting their standard routine care.
4. Results
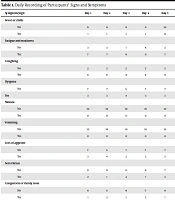
A total of 10 patients met the inclusion criteria to enter the study. There were 6 (60%) male and 4 (40%) female subjects. The mean age of patients was 37.40 ± 7.975 years (minimum 28 years, maximum: 51 years). The mean days passed from the onset of symptoms and referring to Kowsar Hospital in Semnan, Iran, was 3.2 ± 0.919 days (minimum: 2 days, maximum: 5 days). On admission, fever or chills was present in 1 (10%), fatigue and weakness in 7 (70%), dry cough in 8 (80%), dyspnea in 3 (30%), nausea and vomiting in 0 (0%), loss of appetite in 3 (30%), sore throat in 2 (20%), congestion or runny nose in 1 (10%), myalgia in 5 (50%), headache in 2 (20%), diarrhea in 0 (0%), vertigo in 0 (0%), chest pain in 1 (10%), loss of smell in 3 (30%), and loss of taste in 2 (20%) patients. Table 1 shows a daily recording of the participants’ signs and symptoms. Moreover, the mean WBC count on admission was 4.72 ± 0.45 cells/µL. Table 2 shows other laboratory results on admission. Table 3 shows the chest CT scan findings of the patients, such as ground-glass opacities and nodules.
| Symptom/Sign | Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | Day 8 | Day 9 | Day 10 | Day 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Fever or chills | |||||||||||
| No | 9 | 9 | 9 | 9 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| Yes | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Fatigue and weakness | |||||||||||
| No | 3 | 3 | 1 | 4 | 3 | 3 | 4 | 8 | 8 | 9 | 10 |
| Yes | 7 | 7 | 9 | 6 | 7 | 7 | 6 | 2 | 2 | 1 | 0 |
| Coughing | |||||||||||
| No | 2 | 2 | 2 | 2 | 2 | 2 | 4 | 5 | 6 | 7 | 10 |
| Yes | 8 | 8 | 8 | 8 | 8 | 8 | 6 | 5 | 4 | 3 | 0 |
| Dyspnea | |||||||||||
| No | 7 | 7 | 6 | 7 | 7 | 8 | 9 | 9 | 9 | 10 | 10 |
| Yes | 3 | 3 | 4 | 3 | 3 | 2 | 1 | 1 | 1 | 0 | 0 |
| Nausea | |||||||||||
| No | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| Yes | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Vomiting | |||||||||||
| No | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| Yes | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Loss of appetite | |||||||||||
| No | 7 | 6 | 7 | 7 | 7 | 5 | 7 | 8 | 9 | 10 | 10 |
| Yes | 3 | 4 | 3 | 3 | 3 | 5 | 3 | 2 | 1 | 0 | 0 |
| Sore throat | |||||||||||
| No | 8 | 9 | 6 | 9 | 7 | 10 | 10 | 10 | 10 | 10 | 10 |
| Yes | 2 | 1 | 4 | 1 | 3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Congestion or runny nose | |||||||||||
| No | 9 | 8 | 9 | 7 | 9 | 10 | 10 | 10 | 9 | 9 | 10 |
| Yes | 1 | 2 | 1 | 3 | 1 | 0 | 0 | 0 | 1 | 1 | 0 |
| Myalgia (muscle or body aches) | |||||||||||
| No | 5 | 6 | 5 | 3 | 6 | 4 | 7 | 8 | 7 | 9 | 10 |
| Yes | 5 | 4 | 5 | 7 | 4 | 6 | 3 | 2 | 3 | 1 | 0 |
| Headache | |||||||||||
| No | 8 | 8 | 7 | 8 | 7 | 6 | 9 | 8 | 10 | 9 | 10 |
| Yes | 2 | 2 | 3 | 2 | 3 | 4 | 1 | 2 | 0 | 1 | 0 |
| Diarrhea | |||||||||||
| No | 10 | 10 | 10 | 10 | 9 | 10 | 10 | 10 | 10 | 10 | 10 |
| Yes | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Vertigo | |||||||||||
| No | 10 | 10 | 10 | 10 | 9 | 10 | 9 | 10 | 10 | 10 | 10 |
| Yes | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Chest pain | |||||||||||
| No | 9 | 9 | 8 | 8 | 8 | 9 | 8 | 8 | 9 | 10 | 10 |
| Yes | 1 | 1 | 2 | 2 | 2 | 1 | 2 | 2 | 1 | 0 | 0 |
| Loss of smell | |||||||||||
| No | 7 | 6 | 6 | 5 | 5 | 6 | 8 | 8 | 8 | 10 | 10 |
| Yes | 3 | 4 | 4 | 5 | 5 | 4 | 2 | 2 | 2 | 0 | 0 |
| Loss of taste | |||||||||||
| No | 8 | 8 | 8 | 8 | 9 | 9 | 9 | 9 | 10 | 10 | 10 |
| Yes | 2 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 0 | 0 | 0 |
| Mean | 3.9 | 4.6 | 4.3 | 4.3 | 3.8 | 2.6 | 1.9 | 1.6 | 0.8 | 0.7 | 0 |
Daily Recording of Participants’ Signs and Symptoms
| Parameter | Mean ± SD |
|---|---|
| WBC | 4.72 ± 0.45 |
| Lymphocytes | 32 ± 9 |
| Neutrophil | 62 ± 10 |
| RBC | 4.96 ± 0.41 |
Mean Laboratory Findings of Patients on Admission (N = 10)
| Finding | Frequency | Finding | Frequency |
|---|---|---|---|
| GCO | 7/10 | PE | 0 |
| Nodules | 1/10 | Cyst | 0 |
| Linear | 0 | Tree-in-bud sign | 0 |
| Crazy paving | 0 | White lung | 0 |
| Consolidation | 1/10 | Cavity | 0 |
| Architectural distortion | 0 | - | - |
Chest Computed Tomography Scan Findings of Participants (N = 10)
According to RT-PCR results, the mean infectivity period after symptom relief was 6.9 ± 5.152 days. It means that the patients were considered completely treated for about 7 days after symptom relief. However, the mean period of total infectivity was 16.6 ± 5.73 days. Additionally, IgG and IgM were evaluated 1 month after symptom relief. Accordingly, seven patients had positive results for IgG and negative for IgM; two patients had IgM positive and IgG negative, and one patient had negative results for both antibodies. This study re-evaluated IgG and IgM levels 1 month later in these three patients with negative results for IgG. According to the results, two patients developed a positive result for IgG, despite a negative IgM result, and one patient still remained seronegative even after 2 months of symptom relief. The aforementioned three patients with a negative result for IgG 1 month after symptom relief had a longer period to achieve a negative result in RT-PCR. However, there was no statistically significant association between longer RT-PCR positivity and the duration of signs and symptoms (P < 0.21).
5. Discussion
The emergence of the novel coronavirus from the wet markets in China has changed todays’ life to a great extent and affected numerous aspects of human life. According to the Google homepage, up to August 30, 2020, more than 25 million individuals have been infected, and around 843,000 cases died, according to official reports (24). Human-to-human transmission has been suggested as the most important route of infection (25). As most infected patients are asymptomatic, the majority of the present burden might probably be due to the virus’ transmission by asymptomatic carriers (26, 27). On the other hand, those who have been recently recovered might be a source of infection as most think they are not carriers when the signs and symptoms are relieved (28). Therefore, it is important to be aware of how long patients would transfer the infection to others after symptom relief. To date, RT-PCR has been the only standard test to confirm the diagnosis of SARS-CoV-2 RNA, especially to screen symptomatic cases (29, 30). The assessment of humoral immune responses through the detection of IgM and IgG specific to SARS-CoV-2 has also been recognized for the screening of asymptomatic cases and undetectable viral results of RT-PCR (31).
To the best of our knowledge, there is no consensus regarding the infectivity period after symptom relief in COVID-19 patients. However, the USA Centers for Disease Control and Prevention (CDC) claimed that for patients with mild to moderate COVID-19 infection, the replication-competent virus was not observed after 10 days following the initiation of signs and symptoms (32-36). On the other hand, the replication-competent virus has been reported within 10 - 20 days after symptom onset in some patients with severe COVID-19 infection, which might have been associated with an underlying immunocompromised status (37). Nonetheless, it was reported that 88% and 95% of patients did not yield the replication-competent virus after 10 and 15 days following the initiation of signs and symptoms, respectively. This finding is approximately in line with the findings of the present study; however, the current study showed that the total period of infectivity was about 17 days since the initiation of symptoms, which is longer than what was recommended by the CDC. Although the replication-competent virus could not be detected after 3 weeks of symptom onset, the upper respiratory tract samples in some patients still had an RT-PCR positive result for COVID-19 after 12 weeks (38, 39).
On the other hand, researchers claimed that contact with such recovered patients could not transfer the virus to other individuals and therefore was not a source of transmission (32). This is still under debate why such patients have a longer period of RT-PCR positive results but cannot transfer the infection. It is probably related to the viral load in the nasopharyngeal cavity, which could not be infective for other individuals who are in contact. In addition, some other case reports have reported a persistent RT-PCR positive result up to 18 days of symptom onset, which is partly consistent with the present study results.
Moreover, several studies reported that IgM increases during the first week after SARS-CoV-2 infection (ie, 3 - 10 days after the onset of symptoms), reaches the peak level in 2 - 3 weeks, and then reduces to near-background level in most patients. Likewise, IgG responses are initiated that play a key role in long-term immune memory. Long et al. showed that 100% and around 94% of COVID-19 patients had positive virus-specific IgG and IgM approximately 17 - 19 and 20 - 22 days after symptom onset, respectively (40). Consistently, 70% of the patients in the current study had a positive result for IgG and negative for IgM 1 month after symptom relief. However, some individuals develop IgM/IgG antibodies very late after infection, and it has not yet been clear how long these antibodies can be detected. According to the present study results, 20% of patients revealed a weak antibody response, and 10% showed no detectable antibody response during 2 months after symptom relief. The patients also had a longer period to achieve a negative result in RT-PCR. However, delayed immune response might increase the persistence of the virus in the body, even despite clinical symptom relief.
Cheng et al. tried to evaluate the transmissibility of COVID-19 in close contact. They reported that the attack rate was higher among the 1818 contacts whose exposure to index cases started within 5 days of symptom onset but still could occur in other periods. They indicated that high transmissibility of COVID-19 before and immediately after symptom onset plays an important role in controlling the disease spread but is not enough, and asymptomatic patients’ role should also be considered (41). Moreover, Liu et al. claimed that the virus could be detected in COVID-19 patients after resolving clinical symptoms (42). These questions are very important to answer with regard to following a symptom-based or an RT-PCR-based approach for quarantine and isolation rules because there is still not enough evidence to indicate the exact infectivity period of COVID-19 infection after symptom relief. There is a recent trend by newly published investigations to put aside a re-test RT-PCR to show the end of the infectivity period, although there is a long way to reach a consensus in this regard.
As the patients in this study were quarantined up to the end of a positive result in RT-PCR, it was not possible to assess the probability of viral transmission to others. This was due to a lack of enough knowledge in this regard and ethical issues. However, this study did not measure the viral load quantitatively, which could be considered a limitation in this trial. Finally, the present study showed that the virus could be detected in the nasopharyngeal and oropharyngeal samples of the patients up to 7 days after symptom relief. This result highlights the importance of isolation and distancing in patients after recovery and the adoption of hygiene measures. However, this study did not assess whether the patients could infect others in this 7-day period, which was out of control due to insufficient knowledge and ethical issues.
.jpg)