1. Background
Numerous people suffer from food poisoning every year. Staphylococcus aureus is one of the most common causes of food poisoning, which imposes a significant economic burden. Food poisoning with this bacterium is caused by the enterotoxigenic strains in foodstuff (1). It is characterized by a short incubation period, typically 2 - 4 h. Nausea, vomiting, stomach cramps, retching, and prostration are the predominant symptoms; although diarrhea is also often reported, recovery is typically complete within 1 - 2 days. Staphylococcal intoxication is the second most common cause of food poisoning, which often occurs through eating out during travels, hospitalization, etc. Exposure to staphylococcal enterotoxins (SEs) usually does not result in significant mortality, but it can cause illness in 80% of cases that require medical attention and support (2-4). These enterotoxins are extracellular proteins with a molecular weight of 22 - 29 kDa that are similar in composition and biological activities but differ in antigens. They are classified based on their biological and serological characteristics. More than 95% of SEs that cause food poisoning are among the classic SEs, including SEE, SEC, SED, SEA, and SEB. These toxins are resistant to heat and have unique physical and chemical properties, maintaining their biological activity in food even after heat treatment (5-7) in individuals. It particularly resides in the nasal tract, where is found in 20 - 50% of healthy individuals. It can be isolated from hands, feces, and skin. Therefore, people who work in food preparation, processing, and distribution centers can transmit the bacteria to foodstuff in unsanitary conditions. In addition, such individuals can act as dangerous reservoirs of antibiotic-resistant strains of S. aureus, as the misuse/overuse of antimicrobials in the community has led to a significant rise in the prevalence of antibiotic-resistant, coagulase-positive staphylococci. These bacteria contain antibiotic resistance-encoding genes, which are usually on the mobile genetic elements and allow them to be transmitted horizontally to other pathogenic staphylococci and even other bacteria. The spread of resistance genes might lead to life-threatening illnesses (8, 9). Standard immunological methods can be used for the identification of SEs, but these methods have shortcomings, such as a long preparation process, cross-reactivity, and the possibility of false responses. Polymerase chain reaction (PCR) can detect enterotoxin-producing strains, especially when the enterotoxin genes are not expressed for any reason. This technique has been developed using primers directed against sequences in the S. aureus genome, including the thermostable nuclease and enterotoxins.
2. Objectives
Food handlers are frequently contaminated with SEs; even in small amounts, this can lead to severe food poisoning (10). Therefore, the present study is designed to detect enterotoxins A-E of coagulase-positive staphylococci in restaurant employees.
3. Methods
3.1. Specimen Collection and Bacterial Screening
In this experimental study, samples were taken from 91 employees working at 14 restaurants in the Golestan Province (Iran) during 2020 - 2021. The samples were collected from the anterior nares and hands (palms, wrists, fingers) (5) using a cotton swab soaked in sterile saline and then cultured in mannitol salt agar (Merck, Germany). After incubation at 37°C, mannitol-positive S. aureus strains were identified by examining colony morphology, Gram staining, hemolysis, catalase, coagulation, and DNase tests, as well as genotypic analysis. For PCR genotyping, DNA was extracted by the boiling method. For this purpose, a few colonies of S. aureus were incubated for 12 hours in 1 ml of BHI agar. After centrifuge at 3500g rpm for 10 minutes, the supernatants were completely drained, and 800 microliters of lysing buffer were added to the sample. It was incubated at 65oc for 30 minutes in Ben Mari. Specific primers for S. aureus genomic DNA (forward: 5'-AAAAACACTTGTCGATATGG-3 '; reverse: 5'-GTTTCAATACATCAACTGC-3') were designed. Finally, PCR products were electrophoresed on 1.5% agarose gel and visualized under ultraviolet light. Detection of a fragment sized 950 bp confirmed the presence of S. aureus (Figure 1).
3.2. Antibiotic Susceptibility Test
Antibiotic susceptibility was assessed by the disk diffusion method (Kirby-Bauer method). For this purpose, a 24-hour bacterial suspension of S. aureus isolates with 0.5 McFarland turbidity was prepared in physiological serum and then uniformly cultured onto Mueller–Hinton agar (Merck, Germany) using a sterile swab. Antibiotic disks included cefoxitin (30 µg), teicoplanin (30 µg), vancomycin (5 µg), ciprofloxacin (10 µg), cefazolin (30 µg), clindamycin (2 µg), azithromycin (15 µg), daptomycin (2 µg), and amikacin (3 µg) were purchased from the Padten Teb Company (Iran), and linezolid (10 µg) were purchased from Mast Co. (UK). The disks were placed on the plates containing S. aureus isolates for 18 - 24 hours at 37°C. Finally, the diameter of the growth inhibition zone around each disk was measured. According to the Clinical and Laboratory Standards Institute document M100-S25 (2015), an inhibition growth zone diameter of ≤ 21 mm around 30 µg cefoxitin disk and ≤ 10 mm about 1 µg oxacillin disk indicate methicillin resistance (11). As described by Magiorakos et al., MDR is defined as non-susceptibility to ≥ 1 antimicrobial agent in ≥ 3 antimicrobial categories, and XDR is defined as non-susceptibility to ≥ 1 (9). The standard strain ATCC29213 was used as the control strain.
3.3. Detection of SEs Production Ability
To detect enterotoxin genes in S. aureus isolates, DNA was extracted using a commercial kit (Sinaclon, Iran) according to the manufacturer's instructions. The quality and quantity of the extracted DNA were evaluated by spectrophotometry and electrophoresis on 1% agarose gel, respectively. The PCR reaction was performed with specific primers (Table 1) in Mastercycler gradient thermocycler (Eppendorf, USA) under the following cycling conditions: initial denaturation at 95°C for 3 minutes, one cycle of denaturation at 95°C for 1 minute, annealing at 56 - 60°C for 1 minute, 35 cycles of extension at 72°C for 7 minutes, and one cycle of final extension at 72°C for 3 minutes. Next, the PCR products were electrophoresed on 1% agarose gel (12).
| Gene | Sequence (5`>3`) | Size (bp) | Product Size (bp) |
|---|---|---|---|
| sea | F: GGTTATCAATGTGCGGGTGG | 20 | 102 |
| R: CGCCACTTTTTTCTCTTCGG | 20 | ||
| seb | F: GGTGGTGTAACTGAGC | 21 | 164 |
| R: CAAATAGTGACGAGTTAGG | 20 | ||
| sed | F: CCAATAATAGGAGAAAATAAAAG | 23 | 378 |
| R: ATTGGTATTTTTTTTCGTTC | 20 | ||
| see | F: AGGTTTTTTCACAGGTCATCC | 21 | 209 |
| R: CTTTTTTTTCTTCGGTCAATC | 21 |
3.4. Statistical Analysis
Data were presented using frequency charts and numerical indicators. Statistical data analysis was conducted using the chi-square test with the SPSS software (version 18). A P-value of less than 0.05 indicated a statistically significant difference.
4. Results
4.1. Results of Bacteriological Assay
Based on the bacteriological examinations, 29 isolates (31.9%) were identified as S. aureus. The frequency of S. aureus isolates was highest in the waiters (65.5%), whose ages ranged from 34 to 49 years (41.4%), and nasal samples (55.1%) (Table 2).
| Variables | Chefs/Master Chefs | Cleaning Staff | Servers | P-Value |
|---|---|---|---|---|
| Gender | 0.06 | |||
| Male | 9 (31) | 4 (13.8) | 6 (20.7) | |
| Female | 4 (13.8) | 3 (10.3) | 3 (10.3) | |
| Age range (y) | 0.10 | |||
| 18 - 33 | 0 | 3 (10.3) | 7 (24.1) | |
| 34 - 49 | 1 (3.5) | 4 (13.8) | 7 (24.1) | |
| 50 - 65 | 2 (6.9) | 3 (10.3) | 2 (6.9) | |
| Specimen | 0.03 | |||
| Nasal | 1 (3.4) | 6 (20.7) | 9 (31) | |
| Wrist | 0 | 1 (3.5) | 4 (13.8) | |
| In-between the fingers | 1 (3.5) | 3 (10.3) | 4 (13.8) |
a Values are expressed as No. (%).
4.2. Results of the Detection of SEs Gene
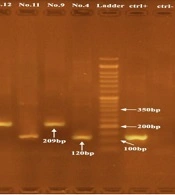
The presence of 120, 164, 209, and 378 bp fragments indicated the presence of the sea, seb, see and sed genes, respectively. Out of 29 S. aureus isolates, the enterotoxin-encoding genes were found in seven samples (24.1%). The most prevalent enterotoxin was SEA (57.1%), while SED and SEE were present only in two strains (28.6%) and one strain (14.3%), respectively. None of the isolates had the seb gene (Figure 2).
4.3. Data on Drug-Resistance Frequency
The highest and lowest susceptibility rates were against linezolid (100%) and cefoxitin (17.24%), respectively (Table 3). Among 19 methicillin-resistant S. aureus (MRSA) isolates, 17 (89.47%) were multidrug-resistant (MDR), and two strains (10.52%) were extensively drug-resistant (XDR). The MRSA isolates were mainly susceptible to linezolid (100%) and vancomycin (48.27%). Out of the seven isolates containing the enterotoxin-encoding genes, five isolates were MDR.
| Susceptibility Result | Frequency of S. aureus Isolates | Chi-Squared (χ2) |
|---|---|---|
| Cefoxitin | 9.77 b | |
| R | 19 (65.51) | |
| S | 10 (34.48) | |
| Teicoplanin | 11.67 b | |
| R | 6 (20.68) | |
| S | 14 (48.27) | |
| Vancomycin | 8.32 b | |
| R | 5 (17.24) | |
| S | 14 (48.27) | |
| Cefazolin | 8.87 b | |
| R | 5 (17.24) | |
| S | 15 (51.72) | |
| Azithromycin | 10.98 b | |
| R | 7 (24.13) | |
| S | 14 (48.27) | |
| Amikacin | 7.87 | |
| R | 11 (37.93) | |
| S | 12 (41.37) | |
| Daptomycin | 11.98 b | |
| R | 6 (20.68) | |
| S | 14 (48.27) | |
| Ciprofloxacin | 9.88 | |
| R | 9 (31.03) | |
| S | 9 (31.03) | |
| Linezolid | 11.76 b | |
| R | 0 (0) | |
| S | 29 (100) | |
| Clindamycin | 11.02 b | |
| R | 13 (44.82) | |
| S | 7 (24.13) | |
| Chi-square (χ2) | 11.74% b |
Abbreviations: R, resistant; S, sensitive..
a Values are expressed as No. (%) unless otherwise indicated.
b Statistically significant difference (P < 0.05).
5. Discussion
Staphylococcal enterotoxin-poisoning usually has a short incubation period, and the symptoms such as nausea, vomiting, muscle, abdominal pain, and diarrhea appear after ingestion. The amount of S. aureus in food is related to several factors, such as the number of bacterial carriers involved in food preparation, sanitation and hygiene of the food factories, and the transportation system (13). In the present study, out of 91 samples taken from restaurant staff, S. aureus was isolated from 29 samples (32%), while 55% of the subjects were nasal carriers of S. aureus. Previous studies in Iran in 2011 (14) and 2018 (15) reported that the frequency of S. aureus carriers was 32% and 26.8% among the hospital staff, respectively. In line with our findings, studies in the Netherlands (16) and Brazil (17) reported that 20-50% and 22.1% of adults were nasal carriers of S. aureus, respectively. In a study in 2010, S. aureus was detected in 57.3% of dairy samples (18). In Germany, a study identified S. aureus in 69 of 135 red meat samples taken during different ham production stages (19). Unlike the mentioned studies, we investigated the frequency of S. aureus isolates in people involved in food production and supply.
Staphylococcal enterotoxins are the main causes of food poisoning that are expressed and transmitted through mobile genetic elements, plasmids, chromosomes, and bacteriophages (7). In this study, among 29 S. aureus isolates, seven isolates contained enterotoxin-producing genes (enterotoxin A-E) except for seb. In addition, none of the isolates had more than one gene. In a previous study, out of 132 S. aureus isolates from dairy products, 90 isolates (68.18%) had one or more enterotoxin-encoding genes (12). In a study in northern Palestine, 37% of S. aureus isolates from dairy products contained enterotoxin-encoding genes, but none had more than one gene (20). The discrepancy in the frequency of enterotoxin-encoding genes in different studies may be due to the differences in the source of bacteria isolation, study location, bacterial detection methods, the number of samples, and the type of samples studied. Moreover, the lower frequency of enterotoxin-producing S. aureus strains in the present study could also be due to the coronavirus disease 2019 pandemic and the restaurant staff's mandatory use of facial masks. In this study, the most and the least prevalent enterotoxin genes were sea and see, respectively, which is in agreement with the results of a study in Poland (20). Similarly, a study in Turkey also reported sea as the most abundant enterotoxigenic gene in foodstuff (21). In the UK, type A is responsible for 52% of outbreaks, type D for 6%, types A and D combined for 19%, and types C and D combined for 9% (22).
In addition to tracking infectious agents and their metabolites, the investigation of bacterial drug resistance always has interested researchers. The prevalence of infections caused by S. aureus is increasing, and the bacterium's ability to acquire and spread multi-drug resistance further highlights the importance of investigating its prevalence in different communities. In recent years, vancomycin has been regarded as a highly effective antibiotic in eliminating Gram-positive bacteria. Still, its overuse has increased the rate of resistance to this antibiotic (23-25). In this regard, 48% of the S. aureus isolates in our study were resistant to vancomycin. Although S. aureus isolates were susceptible to linezolid, the high rate of resistance to commonly-used antibiotics such as teicoplanin, azithromycin, and vancomycin was relatively high. In addition, 89.47% of the isolates were identified as MDR. The frequency of MDR and MRSA isolates in our study was similar to that in other studies (26-28). Still, considering that these isolates were taken from carriers, not patients, detecting 85% of MDR strains, that 29.4% of them are enterotoxigenic, is alarming.
5.1. Conclusions
The results showed that people involved in preparing and supplying food might be a potential source of MDR, enterotoxigenic strains of S. aureus. The relatively high prevalence of enterotoxin-encoding genes, particularly sea and sed, in the isolates indicate the potential risk of food poisoning after eating at the studied restaurants. Further research is needed to screen people involved in food production and supply as main reservoirs of staphylococcal strains.