1. Background
Odontogenic keratocysts (OKCs) are distinct cystic lesions of the jaw characterized by their aggressive nature, high recurrence rates, and potential for malignant transformation (1, 2). Accurate diagnosis of OKCs is critical, as they often share overlapping histopathological features with other odontogenic cysts, such as dentigerous and radicular cysts, making differentiation challenging (1, 3). Misdiagnosis can lead to suboptimal treatment strategies, resulting in higher recurrence rates, increased surgical interventions, and prolonged patient morbidity. Given these clinical implications, there is a pressing need for more precise and reliable diagnostic approaches (1, 4).
Traditional diagnostic methods rely on manual examination of histopathologic slides, which, although considered the gold standard, have inherent limitations. These include interobserver variability, subjectivity, and diagnostic inconsistencies, particularly in cases with subtle morphological differences (5-10). Moreover, manual evaluation can be time-consuming and labor-intensive, further complicating timely and accurate diagnoses in high-volume pathology settings. Delays in diagnosis can negatively impact treatment planning, potentially leading to disease progression and an increased healthcare burden (5, 7, 10-13). These limitations underscore the need for more objective, efficient, and reproducible diagnostic tools.
Artificial intelligence (AI), particularly machine learning (ML) techniques, has emerged as a transformative solution for overcoming these challenges (14-16). Deep learning models, including convolutional neural networks (CNNs), have demonstrated exceptional capabilities in analyzing medical imaging data, including histopathologic slides (17-19). Unlike traditional methods, AI-driven models can rapidly process large datasets, identify complex histopathological patterns with high precision, and minimize diagnostic discrepancies among pathologists. These models have also shown the potential to enhance reproducibility, reduce diagnostic turnaround time, and improve workflow efficiency, addressing key limitations of conventional approaches (19-21).
Notably, deep learning models can detect subtle histological features that may be overlooked by human observers, resulting in superior diagnostic accuracy. Additionally, AI-driven analysis significantly accelerates the diagnostic workflow by processing thousands of images in a fraction of the time required for manual examination, making it a valuable tool for high-throughput pathology applications. Successes in various medical fields, such as cancer detection, dermatopathology, and gastrointestinal pathology, further underscore the potential of AI to improve diagnostic accuracy and efficiency (19, 20, 22). However, the application of AI to odontogenic lesions, including OKCs, has faced both successes and limitations. While AI-based approaches have shown promise for diagnosing odontogenic lesions in radiographic images, their performance in analyzing histopathologic slides has been less extensively investigated and remains an area requiring further exploration.
Several studies have applied AI to the diagnosis of odontogenic lesions, with varying degrees of success (21, 23-26). For example, AI has achieved high diagnostic accuracy in detecting odontogenic lesions in radiographic and cone beam computed tomographic (CBCT) imaging. In the study by Fedato Tobias et al. (27), AI models demonstrated high sensitivity and specificity in detecting and classifying lesions such as OKCs, ameloblastomas, and dentigerous cysts from radiographic images. Similarly, the study by Shrivastava et al. (28) yielded comparable results through the analysis of radiographic and CBCT images.
However, the application of AI to the analysis of histopathologic images of OKCs has been less successful, largely due to the scarcity of high-quality annotated datasets. The limited availability of diverse and well-curated histopathologic images restricts model generalizability and increases the risk of overfitting, particularly when deep learning models are trained on small sample sizes. Inconsistent staining protocols, imaging artifacts, and variations in histological interpretation further contribute to these challenges, making it difficult to develop robust AI models that perform consistently across different datasets (21, 26, 27).
To address these challenges, techniques such as data augmentation, transfer learning, and generative adversarial networks (GANs) have been explored to improve model robustness (15, 19, 29, 30). Data augmentation artificially expands training datasets by applying transformations such as rotation, flipping, and contrast adjustments, thereby enhancing model adaptability to real-world variations. Transfer learning enables AI models to leverage pre-trained knowledge from larger medical imaging datasets, reducing the reliance on extensive OKC-specific data. Generative adversarial networks have also been utilized to generate synthetic histopathologic images, providing additional training data to enhance model performance. Implementing these strategies may help mitigate the limitations posed by small sample sizes, improve generalizability, and increase diagnostic accuracy (19, 30-32). Moreover, AI models can be trained not only to diagnose OKCs but also to assess prognostic markers, enabling clinicians to predict recurrence risk and tailor personalized treatment plans. However, while AI has been used to predict recurrence risks in some odontogenic lesions, studies specifically addressing its prognostic capabilities for OKCs remain scarce, highlighting an important gap in current research (33, 34). Understanding AI’s role in both diagnosis and prognosis is essential for integrating these models into clinical workflows and ultimately improving patient outcomes.
This systematic review and meta-analysis aim to address this gap by evaluating the diagnostic and prognostic performance of AI models in detecting OKCs from histopathologic images. While previous systematic reviews, including recent publications by Fedato Tobias et al. (27) and Shrivastava et al. (28), have explored the use of AI in diagnosing odontogenic lesions, their focus has primarily been on radiographic and CBCT imaging.
This study differs significantly by providing a comprehensive analysis of AI’s diagnostic and prognostic performance in histopathologic image analysis — an area that has received comparatively less attention. By synthesizing data from previous studies, this work aims to provide a critical assessment of the current state of AI applications in this domain, highlight existing limitations, and propose directions for future research and clinical implementation. Ultimately, this review seeks to bridge the gap between AI research and clinical practice, offering valuable insights for pathologists, researchers, and healthcare professionals interested in integrating AI into odontogenic pathology.
2. Methods
2.1. Protocol and Registration
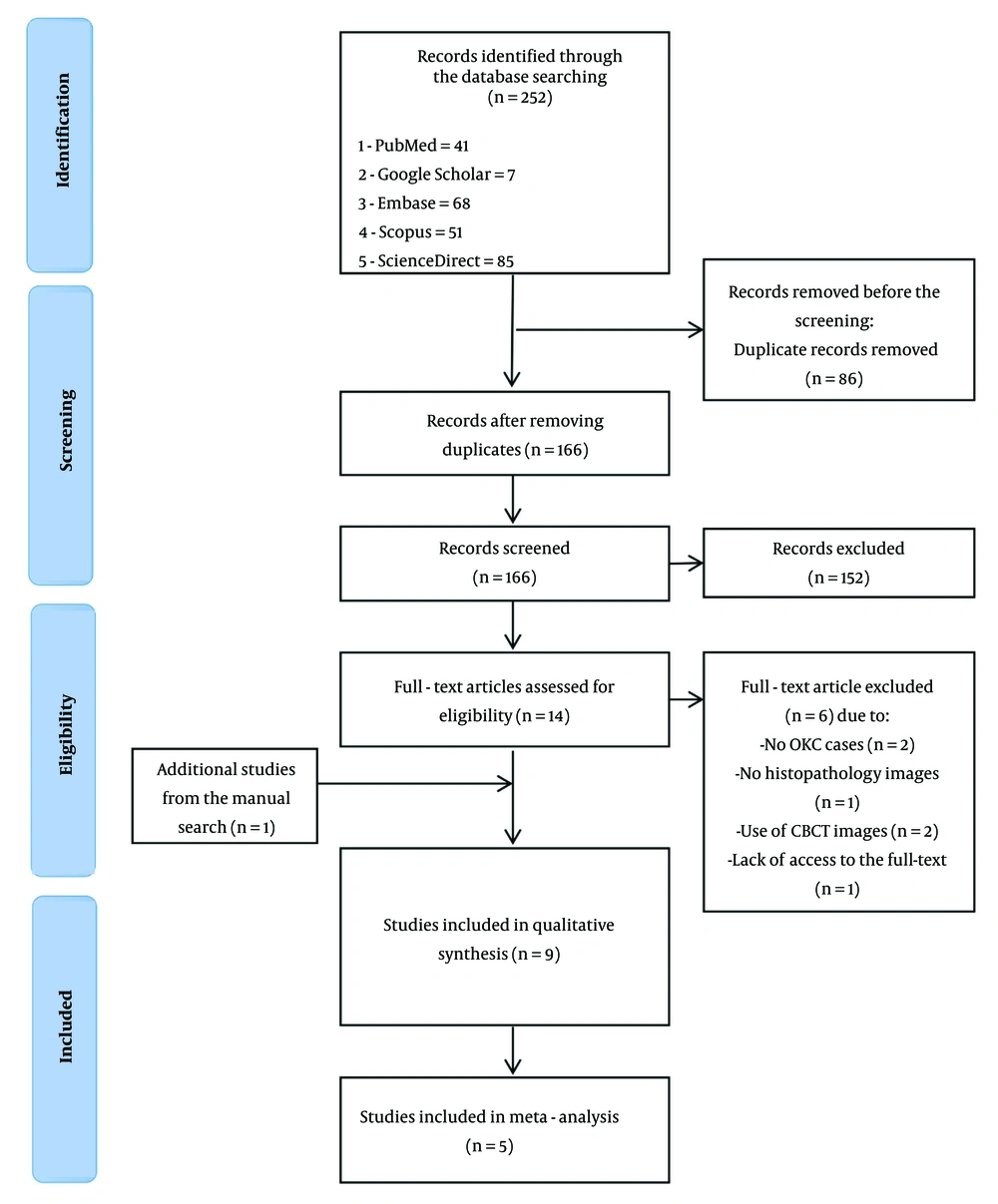
The preferred reporting items for systematic reviews and meta-analyses (PRISMA) flow diagram used to track the study selection process is presented in Figure 1. This systematic review adheres to the guidelines outlined in the PRISMA, specifically the extension for diagnostic test accuracy studies (PRISMA-DTA) (35). The protocol for this review has been registered with the international prospective register of systematic reviews (PROSPERO) under the registration number CRD42024607673.
2.2. Search Strategy
The search strategy for this systematic review involved conducting a comprehensive literature search across PubMed, Scopus, Embase, Google Scholar, and ScienceDirect, covering publications from their inception to May 5, 2024, with an update performed on January 9, 2025. The selection of search terms was based on a combination of the study’s keywords, corresponding medical subject headings (MeSH) terms in PubMed, and insights drawn from existing AI-based systematic reviews in related fields. Additionally, expert opinion was sought to optimize and refine the keyword list. Several key strategies were employed to ensure comprehensive coverage of relevant studies. Table 1 provides detailed information on the search queries used for each database.
| Dataset | Search Query | Results |
|---|---|---|
| PubMed | ("artificial intelligence" OR "artificial intelligence"[MeSH Terms] OR "deep learning" OR "deep learning"[MeSH Terms] OR "machine learning" OR "machine learning"[MeSH Terms] OR “convolutional neural networks” OR “convolutional neural networks"[MeSH Terms]) AND (“odontogenic keratocyst” OR “odontogenic keratocyst”[MeSH Terms] OR “jaw cysts” OR “jaw cysts”[MeSH Terms] OR “Odontogenic cysts” OR “Odontogenic cysts”[MeSH Terms]) | 41 |
| Scopus | TITLE-ABS-KEY (["artificial intelligence" OR "deep learning" OR "machine learning" OR "convolutional neural networks"] AND ["odontogenic keratocyst" OR "jaw cysts" OR "Odontogenic cysts"]) | 51 |
| ScienceDirect | ("artificial intelligence" OR "deep learning" OR "machine learning" OR "convolutional neural networks") AND (“odontogenic keratocyst” OR “jaw cysts” OR “Odontogenic cysts”) | 85 |
| Embase | ('artificial intelligence'/exp OR 'artificial intelligence' OR 'deep learning'/exp OR 'deep learning' OR 'machine learning'/exp OR 'machine learning' OR 'convolutional neural networks') AND ('odontogenic keratocyst'/exp OR 'odontogenic keratocyst' OR 'jaw cysts'/exp OR 'jaw cysts' OR 'odontogenic cysts'/exp OR 'odontogenic cysts') | 68 |
| Google Scholar | Allintitle: ("artificial intelligence" OR "deep learning" OR "machine learning" OR "convolutional neural networks") AND (“odontogenic keratocyst” OR “jaw cysts” OR “Odontogenic cysts”) | 7 |
No language or time restrictions were applied during the search. Additionally, the reference lists of the included articles were screened to identify any additional relevant studies.
2.3. Inclusion and Exclusion Criteria
Studies were eligible for inclusion if they addressed the PICO (patient/population, intervention, comparison, and outcomes) framework by evaluating whether AI models (I) can enhance diagnostic and prognostic accuracy (O) for OKCs in histopathologic images (P): Population (P): Histopathologic images of OKCs; intervention (I): Use of AI or MI models for diagnostic purposes; comparison and outcomes (C, O): Studies were required to report at least one performance metric, such as accuracy, sensitivity, specificity, or area under the curve (AUC); study design: This review included original research articles employing experimental, observational, retrospective, or prospective study designs.
Case reports, reviews, editorials, and commentaries were excluded. Additionally, studies that did not report diagnostic performance metrics or used only panoramic or CBCT images for diagnosis were excluded.
2.4. Study Selection
EndNote X9 was used to manage citations. After eliminating duplicates, three reviewers independently screened the titles and abstracts of the identified studies. Full-text articles were retrieved for studies deemed potentially eligible by the lead reviewer. Any discrepancies among the three reviewers were resolved through discussion, and, if necessary, with the input of a fourth reviewer.
2.5. Data Extraction
Two trained researchers and dentists independently extracted data from the included studies using a predefined data extraction form. The extracted data were verified by two reviewers to reduce the risk of any possible bias. The collected data include study characteristics such as author, year, sample size, inclusion and exclusion criteria, artificial intelligence model (types of models like convolutional neural networks, support vector machines), architecture, training, and testing datasets, diagnostic performance including sensitivity, specificity, accuracy, and area under the curve, data preprocessing methods such as data augmentation and image normalization.
Additionally, we recorded details regarding dataset size, the number of histopathologic images used for training and validation, and whether studies employed single-institution or multi-institutional data sources. Studies that did not specify dataset characteristics or validation procedures were noted. Discrepancies in data extraction were resolved through consultation with the reviewer.
2.6. Quality Assessment
The risk of bias in the included studies was independently assessed by three reviewers using the quality assessment of diagnostic accuracy studies tool. This tool evaluates four domains: patient selection, index test, reference standard, and flow and timing. Each study was rated as having a low, high, or unclear risk of bias in each domain.
To specifically assess the reliability of the datasets used in artificial intelligence model training, we examined whether studies reported dataset size, data augmentation techniques, external validation, and measures to prevent overfitting, such as cross-validation and transfer learning. Studies with small, single-institution datasets or unclear validation procedures were flagged as having potential concerns regarding generalizability.
Any disagreements among the reviewers were resolved through discussion led by the lead reviewer.
2.7. Data Synthesis
The primary outcome measured was the AUC, while secondary outcomes included sensitivity, specificity, and accuracy. A random-effects model was employed to account for heterogeneity among the studies, which was assessed using the I² statistic. The pooled diagnostic accuracy was represented as a summary receiver operating characteristic (sROC) curve. In this meta-analysis, the weight of each study was determined based on its sample size, with larger studies contributing more to the pooled estimates.
Publication bias was evaluated using Egger’s test. All statistical analyses were conducted using Python, employing the NumPy package for data manipulation, Matplotlib for visualization, and scikit-learn (sklearn) for ROC curve analysis. A P-value of less than 0.05 was considered statistically significant.
3. Results
The key findings from the studies are summarized in Table 2.
| Authors (Year) | Study’s Aim | Sample Prevalence | Data Sources | Data Set (Training, Validation, Testing) | Inclusion and Exclusion Criteria | Labeling Procedure | AI Model Task | Pre-processing (P) Augmentation (A) | Model (Algorithm, Architecture) | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| Cai et al. (2024) (5) | Develop AI models for diagnosis and prognosis of OKC | 519 cases (OKC: 400, OOC: 90, GC: 29) | Single-institution | Diagnostic: Training (70% - 363 cases), testing (30% - 156 cases) prognostic: 1688 WSIs, training (280 cases), testing (120 cases) | Excluded cases with unclear or faded H&E staining | NA | Classification | P: Macenko method (tiles of 512 × 512 pixels from WSIs, white background removal using AI, color normalization), Z-score normalization A: Random horizontal and vertical flipping | SVM, random forest, extra trees, XGBoost, LightGBM, MLP | Diagnostic AUC = 0.935 (95% CI: 0.898-0.973) prognostic AUC = 0.840 (95% CI: 0.751-0.930) prognostic accuracy = 67.5%, sensitivity = 92.9% (multi-slide) |
| Kim et al. (2024) (36) | Evaluate the agreement between clinical diagnoses and histopathological outcomes for OKCs and odontogenic tumors by clinicians, ChatGPT-4, and ORAD | 623 specimens (OKC: 321, odontogenic tumors: 302) | Single-institution | NA | Excluded non-odontogenic, metastatic, adjacent spread, and non-OKC cysts | NA | Classification | NA | ChatGPT-4 (language model) ORAD (Bayesian algorithm) | ChatGPT-4 concordance rates: 41.4% ORAD concordance rates: 45.6% |
| Mohanty et al. (2023) (7) | Automate risk stratification of OKC using WSIs | 48 WSIs (508 tiled images) | Multi-institutional | Training (70%), validation (10%), testing (20%) | Excluded blurry images and those with poor staining quality | Manual labeling by pathologists | Classification | P: Entropy/variance calculation to remove white tiles A: Image rotation, flipping | Attention-based image sequence analyzer (ABISA), Vision Transformer, LSTM | ABISA: Accuracy = 98%, Sensitivity = 100%, AUC = 0.98 VGG16: Accuracy = 80%, AUC = 0.82 VGG19: Accuracy = 73%, AUC = 0.77 inception V3: Accuracy = 82%, AUC = 0.91 |
| Mohanty et al. (2023) (6) | Build automation pipeline for diagnostic classification of OKCs and non-KCs using WSIs | 48 OKC slides, 20 DC slides, 37 RC slides, 6069 OKC tiles, 5967 non-KC tiles | Multi-institutional | Training (80%), validation (20%) | Excluded blurry tiles, white tiles, and those with non-important information using OTSU thresholding | Manual labeling by pathologists | Classification | P: Tile generation (2048 × 2048), white tile removal, OTSU thresholding A: Rotation, shifting, zoom, flipping | P-C-reliefF model (PCA + reliefF), VGG16, VGG19, inception V3, standard CNN | P-C-reliefF: Accuracy = 97%, AUC = 0.99 VGG16: Accuracy = 97%, AUC = 0.93 VGG19: Accuracy = 96%, AUC = 0.93 standard CNN: Accuracy = 96%, AUC = 0.93 |
| Rao et al. (2022) (11) | Develop ensemble deep-learning-based prognostic and prediction algorithm for OKC recurrence | 1660 digital slide images (1216 non-recurring, 444 recurring OKC) | Single-institution | Training (70%), testing (15%), validation (remaining) | Sporadic OKC with 5-year follow-up; excluded syndromic OKC and radical treatment cases | Labeling based on histopathological features (subepithelial hyalinization, incomplete epithelial lining, corrugated surface) | Classification | A: Rotation, shifting, shear, flipping | DenseNet-121, inception-V3, inception-ResNet-V2, novel ensemble model | DenseNet-121: Accuracy = 93%, AUC = 0.9452 inception-V3: Accuracy = 92%, AUC = 0.9653 novel ensemble: Accuracy = 97%, AUC = 0.98 |
| Rao et al. (2021) (37) | Develop a deep learning-based system for diagnosing OKCs and non-OKCs | 2657 images (54 OKCs, 23 DCs, 20 RCs) | Single-institution | Training (70%), validation (15%), test (15%) | Excluded inflamed keratocysts and inadequate biopsies | Manual labeling by pathologists | Classification | A: Shear, rotation, zooming, flipping; region-of-interest isolation | VGG16, DenseNet-169; DenseNet-169 trained on full image and epithelium-only dataset | VGG16: Validation accuracy = 89.01%, Test accuracy = 62% DenseNet-169 (Exp II): Validation accuracy = 89.82%, Test accuracy = 91%, AUC = 0.9597 DenseNet-169 (Exp III): Test accuracy = 91%, AUC = 0.9637 Combined model (Exp IV): Accuracy = 93% |
| Florindo et al. (2017) (38) | Classify OKC and radicular cysts | 150 images (65 sporadic OKCs, 40 syndromic OKCs, 45 RCs) | NA | Random 10-fold cross-validation | Excluded images with artifacts or poor quality | Manual labeling by histopathologists | Classification | P: Grey-level conversion, segmentation | Bouligand-Minkowski descriptors + LDA | 72% (OKC vs. RC vs. syndromic OKC) 98% (OKC vs. radicular) 68% (sporadic vs. syndromic OKC) |
| Frydenlund et al. (2014) (39) | Automated classification of four types of odontogenic cysts | 73 images (20 DC, 20 LPC, 20 OKC, 13 GOC) | NA | Training (38 images), validation (37 images), testing (39 images) | Excluded images with poor quality or resolution issues | Manual labeling by pathologists | Classification | P: Color standardization, smoothing, segmentation | SVM, bagging with logistic regression (BLR) | SVM: 83.8% (validation), 92.3% (testing) BLR: 95.4% (testing) |
| Eramian et al. (2011) (40) | Segmentation of epithelium in H&E-stained odontogenic cysts | 38 training images, 35 validation images | NA | Training: 38 images (10 dentigerous cysts, 10 odontogenic keratocysts, 10 lateral periodontal cysts, 8 glandular odontogenic cysts), validation: 35 images (same distribution) | Excluded inflammatory odontogenic cysts | Manually identified epithelium and stroma pixels | Segmentation | P: Luminance and chrominance standardization | Graph-based segmentation | Sensitivity: 91.5% specificity: 85.1% accuracy: 85% |
Abbreviations: AI, artificial intelligence; AUC, area under the curve; ABISA, attention-based image sequence analyzer; CNN, convolutional neural networks; OKC, odontogenic keratocysts; WSIs, whole slide images.
3.1. Study Selection and Inclusion Process
A total of 252 studies were identified through database searches, with one additional record found through manual searching of reference lists. After removing 86 duplicates, 166 studies remained for title and abstract screening. Following this step, 152 studies were excluded based on irrelevance, leaving 14 studies for full-text review. After full-text evaluation, 6 studies were excluded for not meeting the inclusion criteria (e.g., lack of analysis of OKCs, use of non-histopathologic images, or lack of access to full text). This process resulted in the final inclusion of 9 studies (8 from database searches and 1 from manual search) in this systematic review. Among these, 5 studies provided sufficient data to be included in the meta-analysis.
3.2. Study Characteristics and Data Quality
Nine studies published between 2011 and 2024 met the inclusion criteria, demonstrating progressive advancements in the application of AI for OKC analysis (5-7, 11, 36-40).
3.2.1. Sample Prevalence, Data Sources, and Eligibility Criteria
The sample sizes in the studies varied substantially, ranging from 48 whole slide images (WSIs) in Mohanty et al. (7) to over 1,500 images in other studies (11, 37). The prevalence of OKC cases within these datasets ranged from 20 in Frydenlund et al. (39) and Eramian et al. (40) to 400 cases in Cai et al. (5).
Data sources were diverse, including both single-institution and multi-institution datasets, with variations in image resolution, staining protocols, and acquisition equipment. Most studies relied on single-institution datasets (5, 11, 36, 37), while some incorporated multi-institutional data (6, 7). Additionally, three studies did not specify their data sources (38-40), which may impact dataset diversity and overall generalizability.
The eligibility criteria among the included studies varied, but generally ensured that only high-quality images and relevant cases were included for AI-based analysis. Studies commonly excluded cases with unclear or faded hematoxylin and eosin staining, non-odontogenic or metastatic cysts, and images with poor resolution or artifacts.
3.2.2. Data Preprocessing and Labeling Procedure
To enhance model robustness, most studies implemented comprehensive preprocessing pipelines. Common techniques included color normalization (e.g., the Macenko method), white background removal, and tile generation from WSIs. Additionally, data augmentation strategies — such as random flipping, rotation, shifting, zooming, and shear transformations — were routinely applied to expand the effective size and diversity of the training sets. These preprocessing steps were critical for ensuring consistent image quality and mitigating the effects of dataset heterogeneity, thereby contributing to the high diagnostic performance observed.
In most included studies, samples were labeled by experienced pathologists, ensuring high-quality annotations. Labels were assigned based on characteristic histopathological features of OKCs, including subepithelial hyalinization, incomplete epithelial lining, and surface corrugation.
3.2.3. Training Data Set and Training Process of Models
The number of images in the training sets and the training processes varied significantly across the studies. Training datasets ranged from as few as 38 images in the study by Eramian et al. (40) to more than 1,500 images in the studies by Rao et al. (11, 37). Among the deep learning architectures, DenseNet-169 — employed in the studies by Rao et al. (11, 37) — was trained using the Adam optimizer (learning rate = 0.001, batch size = 12 or higher) and incorporated data augmentation techniques such as shear, rotation, and zoom transformations. DenseNet-169 achieved high classification performance, with reported accuracy exceeding 91% and an AUC ranging from 0.9597 to 0.9653 (11, 37).
Similarly, Inception-V3 — used by Mohanty et al. (6) and Cai et al. (5) — demonstrated robust classification performance, with RMSprop as the optimizer and a learning rate of 0.0001 (5, 6). Additionally, Cai et al. (5) optimized the model using fivefold cross-validation and grid-search hyperparameter tuning, with a batch size of 32 and a dropout rate of 0.5 to prevent overfitting. An advanced hybrid deep learning approach, attention-based image sequence analyzer (ABISA), was also evaluated. This model, proposed by Mohanty et al. (7), integrates convolutional and recurrent layers with an attention mechanism for enhanced interpretability. The ABISA was trained using the Adam optimizer with a cyclical learning rate schedule (0.0001 to 0.001) and extensive data augmentation techniques, including contrast adjustment and histogram equalization (7).
Florindo et al. (38) introduced a morphological classification framework using Bouligand-Minkowski fractal descriptors to differentiate OKCs from other cysts. This method, applied to 150 histological images, demonstrated a classification accuracy of 98%, highlighting the potential of fractal-based feature extraction as a complementary approach to deep learning models.
The study by Eramian et al. (40) further contributed by developing an automated epithelial segmentation algorithm using binary graph cuts. The segmentation model was trained on 38 histological images, achieving a mean sensitivity of 91.5% and specificity of 85.1%, with the highest accuracy observed in OKCs (96.1% sensitivity, 98.7% specificity). Their color standardization and edge detection preprocessing steps were crucial in refining AI-driven histopathological segmentation.
3.2.4. Model Diagnostic and Prognostic Performance
Recent studies (2021 - 2024) exhibited superior diagnostic performance, with accuracy rates consistently exceeding 90% (5, 7, 11, 37). The ABISA achieved the highest diagnostic accuracy (98%, AUC = 0.98) (7), while the P-C-ReliefF model demonstrated comparable performance (97% accuracy, AUC = 0.99) (6). Traditional MI approaches showed varying but promising results, with LightGBM and XGBoost achieving AUCs of 0.935 and 0.93, respectively (5). Deep learning architectures, particularly DenseNet-169 and Inception-V3, demonstrated robust performance with AUCs ranging from 0.9597 to 0.9653 (11, 37), while ensemble approaches further improved accuracy up to 97% (11). Notably, two recent studies specifically addressed OKC recurrence prediction, with Rao et al. (11) achieving 97% accuracy (AUC = 0.98) using a novel ensemble model, while Cai et al. (5) reported a prognostic AUC of 0.840 (95% CI: 0.751 - 0.930). Most studies implemented comprehensive preprocessing pipelines, including color normalization, white background removal, and tile generation from WSIs (5-7, 11, 37). Data augmentation strategies, reported in five studies (5-7, 11, 37), predominantly utilized random flipping, rotation, and shear transformations. Early approaches (2011 - 2017) focused on basic classification tasks using traditional MI methods, achieving moderate success (72 - 98% accuracy range) (38-40), while recent studies employed more sophisticated architectures and addressed complex tasks such as recurrence prediction (5, 7, 11, 37). This temporal progression reflects both methodological advancement and improved performance metrics, with modern approaches consistently achieving AUC values above 0.90 and providing more robust diagnostic and prognostic capabilities (5, 7, 11, 37).
Notably, one study incorporated large language models (LLMs) such as Chat-GPT4, demonstrating comparable diagnostic accuracy to clinicians in diagnosing OKCs, with a concordance rate of 41% and a kappa value of 0.14 (36). While Chat-GPT4 showed strengths in processing textual clinical data, it exhibited slightly lower specificity (90.4%) and sensitivity (30.1%) compared to clinicians (specificity: 95.7%, sensitivity: 32.9%).
3.3. Risk of Bias and Applicability
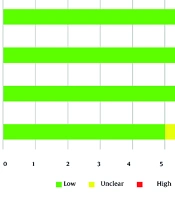
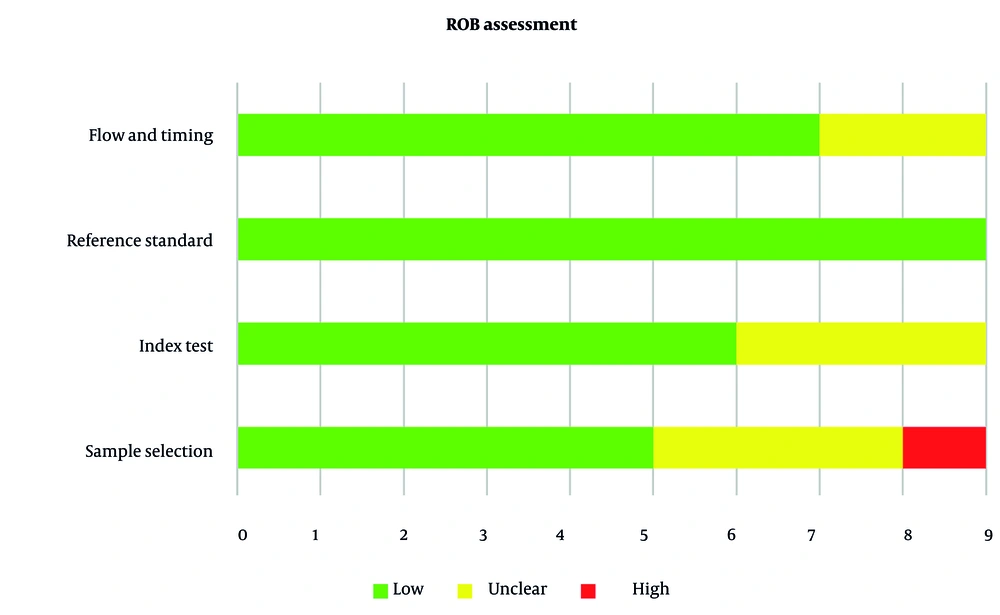
The ROB across the included studies was generally low, with a few exceptions. Four of the nine studies (44.4%) demonstrated low risk across all four domains (5, 7, 11), whereas five studies (55.6%) exhibited concerns in one or more areas. Specifically, in the patient selection domain, 55.6% of studies were rated as low risk, 33.3% as unclear, and 11.1% as high risk. For the index test domain, 66.7% were rated as low risk and 33.3% as unclear, while all studies (100%) showed low risk in the reference standard domain. In the flow and timing domain, 77.8% of studies were classified as low risk, and 22.2% as unclear (Figure 2 and Table 3).
| Authors (Year) | ROB Assessment | Applicability Assessment | |||||
|---|---|---|---|---|---|---|---|
| Patient/Sample Selection | Index Test | Reference Standard | Flow and Timing | Patient/Sample Selection | Index Test | Reference Standard | |
| Cai et al. (2024) (5) | Low | Low | Unclear | Low | Low | Low | Low |
| Kim et al. (2024) (36) | Unclear | Low | Low | Low | Low | Low | Low |
| Mohanty et al. (2023) (7) | Low | Low | Low | Low | Low | Low | Low |
| Mohanty et al. (2023) (6) | Low | Low | Low | Low | Low | Low | Low |
| Rao et al. (2022) (11) | Low | Low | Low | Low | Low | Low | Low |
| Rao et al. (2021) (37) | Low | Unclear | Low | Unclear | Low | Low | Low |
| Florindo et al. (2017) (38) | Unclear | Unclear | Low | Low | Low | Low | Low |
| Frydenlund et al. (2014) (39) | Unclear | Low | Low | Unclear | Low | Low | Low |
| Eramian et al. (2011) (40) | High | Unclear | Low | Low | Low | Low | Low |
3.4. Meta-Analysis
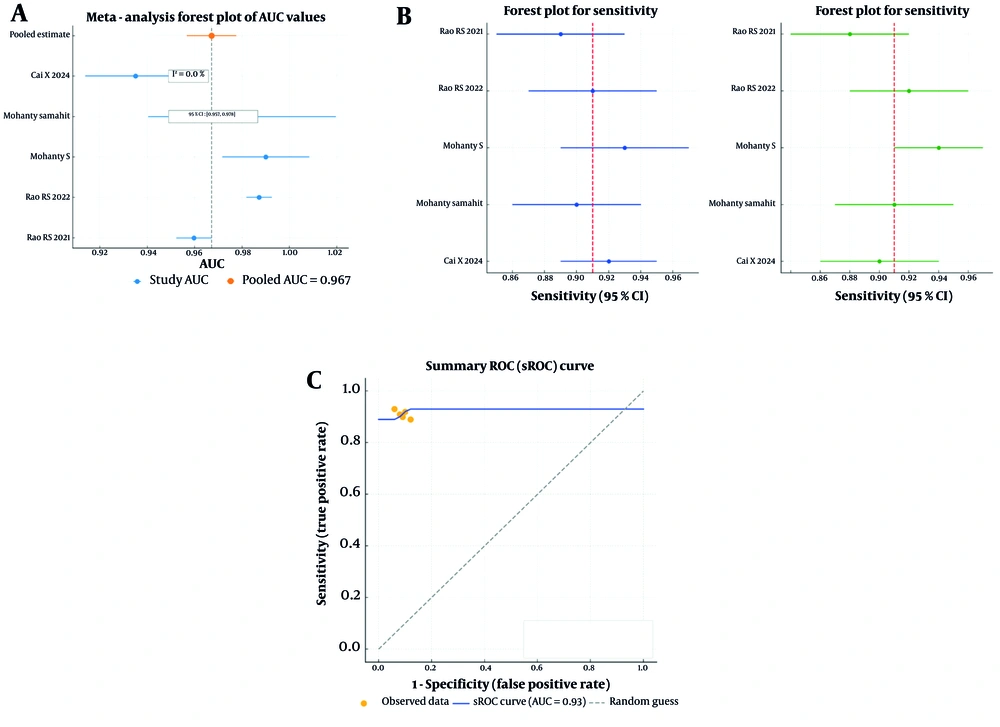
The meta-analysis was conducted to assess the pooled diagnostic accuracy of the included studies. The pooled analysis of five studies that reported the AUC yielded a value of 0.967 (95% CI: 0.957 – 0.978), indicating strong overall diagnostic accuracy. Additionally, heterogeneity among the studies was minimal (I² = 0%), suggesting consistent performance across various AI models and datasets (Figure 3A).
A, The pooled analysis of five studies that reported area under the curve (AUC) with a value of 0.967 (95% CI: 0.957 - 0.978) and low heterogeneity (I2 = 0.0%); B, the pooled sensitivity and specificity across the studies further demonstrated the robust performance of the artificial intelligence (AI) models, the pooled sensitivity was 0.90 (95% CI: 0.89 - 0.92) and the pooled specificity was 0.91 (95% CI: 0.88 - 0.94), the forest plots for both sensitivity and specificity displayed relatively narrow confidence intervals, indicating consistent diagnostic performance across the studies included; C, the summary receiver operating characteristic (sROC) curve had an AUC of approximately 0.93, further highlighting the diagnostic strength of the AI models.
Forest plots for sensitivity and specificity showed that the pooled sensitivity ranged from 0.89 to 0.92, while the pooled specificity ranged from 0.88 to 0.94 (Figure 3B). The sROC curve, which assesses overall diagnostic performance by combining sensitivity and specificity, demonstrated an AUC of 0.93. This result further confirms the high diagnostic accuracy of AI models across studies. Most data points were concentrated near the upper-left corner of the sROC plot, indicating a strong balance between sensitivity and specificity (Figure 3C).
Egger’s test for publication bias produced a P-value of 0.522, suggesting that there was no significant evidence of publication bias.
4. Discussion
This systematic review and meta-analysis explored the diagnostic and prognostic performance of AI models in detecting OKCs from histopathological images. The findings confirm the promising potential of AI — particularly deep learning models — in achieving high diagnostic accuracy, as demonstrated by a pooled AUC of 0.967. The pooled sensitivity of 0.90 suggests that AI systems were effective in identifying positive OKC cases, while a pooled specificity of 0.91 indicates a robust capability to reduce false positives by correctly identifying negative cases. These results emphasize the role AI can play in enhancing clinical decision-making for OKCs, especially in scenarios where access to expert pathologists is limited.
AI models excel at processing intricate histopathological images and autonomously learning hierarchical features (19, 30, 41, 42). Compared to conventional diagnostic methods, AI-driven models significantly reduce interobserver variability and offer a more standardized approach to pathology assessment. Specifically, DenseNet-169, which achieved an impressive AUC of 0.9872, exemplifies the strengths of AI in handling high-resolution medical images due to its efficient feature extraction mechanisms (29, 37, 43). Unlike traditional MI models such as support vector machines (SVMs), deep learning approaches eliminate the need for manual feature extraction, minimizing potential biases and allowing for more adaptive analysis of complex datasets (44-47). This adaptability is particularly beneficial in distinguishing OKCs from histologically similar cystic lesions.
Several studies included in the meta-analysis demonstrated that AI models achieve diagnostic accuracy comparable to, or even surpassing, that of human pathologists (5, 7). The pooled sensitivity (0.89 - 0.92) and specificity (0.88 - 0.94) reflect the high diagnostic precision of AI, which also offers additional benefits such as the ability to process large volumes of data consistently without fatigue (48, 49). Moreover, AI can help mitigate inter-observer variability, a common issue in histopathological evaluations (31, 49-51). This aspect is particularly relevant in diagnosing OKCs, where histopathological features may overlap with those of other odontogenic cysts, leading to potential misclassification.
4.1. Conclusions
In conclusion, AI models represent a significant advancement in diagnosing OKCs, offering high accuracy, sensitivity, and specificity in analyzing histopathologic images. These systems hold great potential for enhancing diagnostic workflows, improving concordance rates, and reducing variability among pathologists. However, several challenges must be addressed to fully realize this potential. A key priority is the external validation of AI models using diverse, large-scale datasets to ensure generalizability across populations. The development of explainable AI tools is equally critical to facilitate trust and adoption among clinicians. Additionally, standardized evaluation methods are essential to enable meaningful cross-study comparisons and to guide clinical integration efforts.
4.2. Comparison with Previous Studies
To the best of our knowledge, no systematic review and meta-analysis have been conducted to date to assess the diagnostic and prognostic performance of AI models in detecting OKCs from histopathologic images. While previous reviews, such as those by Fedato Tobias et al. (27) and Shrivastava et al. (28), have explored the role of AI in diagnosing odontogenic cysts and tumors, their focus was primarily on radiographic and CBCT imaging. In contrast, our systematic review provides a comprehensive evaluation of AI performance in histopathologic image analysis — an area that has received comparatively less attention.
Our findings align with those of Fedato Tobias et al. (27), who reported high sensitivity and specificity of AI models in odontogenic cyst classification. However, our study expands on this by emphasizing the impact of preprocessing techniques, model architecture, and dataset diversity on AI performance. Similarly, Shrivastava et al. (28) highlighted the need for AI validation across multiple imaging modalities, which aligns with our recommendation for external dataset testing. Additionally, Shi et al. (52) focused solely on radiographic images without conducting a meta-analysis or systematic review, further underscoring the novelty and significance of our approach.
4.3. Generalizability and Model Evaluation on External Data
One of the key limitations of current AI models is their limited generalizability due to reliance on single-institution datasets and the lack of external validation. Many of the studies included in this review trained their models on internal datasets (5, 11, 36, 37), raising concerns about their applicability across different populations, imaging settings, and clinical environments.
Potential solutions to these concerns can be considered in future research. Conducting multi-center validation studies will help ensure that AI models generalize well across different clinical settings. Utilizing independent test datasets, instead of relying solely on internal cross-validation, can improve robustness and detect overfitting. Standardizing imaging protocols and annotation methods will reduce inconsistencies in histopathologic image analysis, leading to more reliable AI predictions. Collaborative efforts with leading organizations, including the international organization for standardization (ISO) and the Institute of Electrical and Electronics Engineers (IEEE), can facilitate the creation of these guidelines. Implementing common data models (CDMs) and standardized ontologies, such as the observational medical outcomes partnership (OMOP) and the unified medical language system (UMLS), will harmonize data representation across institutions. Employing domain adaptation techniques can enhance model adaptability to different imaging sources and patient populations. Additionally, federated learning approaches allow AI models to be trained on diverse datasets from multiple institutions without transferring raw patient data, preserving privacy while improving generalizability (11, 37-40).
4.4. Challenges in of Artificial Intelligence-Based Odontogenic Keratocysts Diagnosis
One of the primary objectives of this study was to examine the challenges associated with AI-driven diagnosis, particularly in detecting OKCs. One challenge is the small sample sizes used for training, testing, and validating the model. of Artificial intelligence models trained on datasets with disproportionately low numbers of OKC cases may exhibit bias toward more prevalent cyst types, leading to suboptimal diagnostic performance (53, 54). To mitigate these biases, we recommend using data augmentation, class weighting, and synthetic data generation techniques — such as GANs — to create more balanced datasets (32, 53, 54).
Variability in histopathologic image preprocessing, including color normalization, tile selection, and contrast adjustment, significantly affects AI model accuracy. Inconsistent preprocessing pipelines across studies can lead to performance discrepancies, highlighting the need for standardized methodologies to ensure reproducibility and reliability. Deep learning models often function as "black boxes," limiting the interpretability of their decision-making processes (55-57). Explainability techniques such as Grad-CAM and SHAP values can improve transparency, fostering trust among clinicians and facilitating the integration of AI into diagnostic workflows. Grad-CAM generates heatmaps highlighting the regions of histopathological images that contributed most to the model’s decision, aiding pathologists in understanding why certain areas were classified as OKC. SHAP values, on the other hand, quantify the contribution of each feature in a model’s decision-making process, providing numerical insights into AI predictions. In clinical practice, these techniques can enhance the interpretability of AI-driven diagnoses by offering visual and quantitative explanations, increasing clinician trust, and supporting AI-assisted decision-making (55).
4.5. Practical Applications of Artificial Intelligence Models in Diagnostic Environments
AI has significant potential for integration into real-world diagnostic workflows in hospitals and clinics. By incorporating AI models into pathology laboratories, clinicians can improve diagnostic speed and accuracy, particularly in settings with high case volumes. Potential applications include: (1) Automated screening of histopathologic slides to assist pathologists in prioritizing complex cases; (2) AI-assisted second opinions to reduce diagnostic variability and enhance confidence in challenging cases; and (3) predictive modeling for recurrence risk assessment, enabling personalized treatment planning (15, 28, 52).
For successful integration of AI models into clinical workflows, several structured steps should be considered. First, AI models should be validated on external datasets and multi-center studies to ensure robustness across different clinical environments. Second, AI systems must be seamlessly integrated into existing laboratory information systems (LIS) and digital pathology platforms to facilitate real-time analysis of histopathological slides. Third, AI-driven decision support tools should be designed to provide interpretable outputs, such as probability scores, heatmaps, or case prioritization, to assist rather than replace pathologists in diagnostic decision-making. Fourth, comprehensive training programs should be implemented to familiarize clinicians and laboratory staff with AI-based tools, ensuring their appropriate use and interpretation. Fifth, regulatory approval and compliance with healthcare data protection laws (e.g., HIPAA, GDPR) should be ensured before deployment. Finally, continuous model monitoring and updates based on real-world clinical feedback are essential to maintain accuracy and address evolving diagnostic challenges. These steps will facilitate the responsible and effective implementation of AI in pathology laboratories, ultimately improving diagnostic efficiency and accuracy.
However, practical challenges remain, particularly concerning how AI can be seamlessly incorporated into existing diagnostic systems. To facilitate this, AI tools must be user-friendly and compatible with current diagnostic infrastructure, allowing pathologists to easily interact with and interpret AI results. Additionally, training programs should be developed to familiarize clinicians with the capabilities and limitations of AI tools, ensuring they are used appropriately to augment human expertise.
4.6. Ethical Considerations and Future Recommendations
The integration of AI in pathology necessitates careful ethical consideration to ensure responsible implementation. Adopting the framework proposed by Rokhshad et al. (58), key ethical principles — transparency, fairness, privacy protection, accountability, and equitable access — must guide AI deployment.
4.6.1. Transparency
Transparent AI development, including clear documentation of training data, algorithms, and decision-making processes, is essential to foster trust among clinicians and patients. Active participation from diverse stakeholders, such as pathologists, ethicists, and policymakers, is crucial for maintaining ethical AI applications in pathology (58, 59).
4.6.2. Fairness
Addressing bias in AI models is imperative, as imbalanced training datasets may lead to disparities in diagnostic performance. Ensuring diverse representation in training data and prioritizing equitable access — particularly in low-resource settings — can enhance the fairness of AI applications in pathology (58-60).
4.6.3. Privacy Protection
Given AI’s reliance on sensitive medical data, compliance with data protection regulations (e.g., GDPR, HIPAA) is critical. Techniques like federated learning and anonymization can help maintain privacy while enabling AI models to learn securely from diverse datasets (58).
4.6.4. Accountability and Equitable Access
Artificial intelligence should function as a supportive tool rather than a replacement for human expertise. Clinicians must retain responsibility for final diagnostic decisions, necessitating targeted training programs to improve digital literacy and ensure appropriate AI integration into clinical workflows (58, 59, 61).
AI development should consider long-term sustainability, particularly its environmental impact. Computational efficiency and energy-conscious AI model optimization can mitigate concerns related to high resource consumption and carbon emissions (58, 59, 61).
To facilitate responsible AI adoption, regulatory frameworks should define legal responsibilities for AI developers and healthcare providers. Independent AI oversight committees and interdisciplinary collaborations are essential for standardizing AI integration in diagnostic workflows while addressing ethical concerns (58, 61).
4.7. Environmental Impacts of Developing Artificial Intelligence Models
Deploying AI models in clinical settings offers significant benefits but also raises environmental concerns, primarily due to high energy consumption, increased carbon emissions, and electronic waste. Training sophisticated AI models requires substantial computational power, contributing to rising electricity usage and carbon footprints — particularly if data centers rely on fossil fuels. Additionally, frequent hardware updates generate electronic waste, further impacting the environment. To mitigate these effects, several strategies can be implemented. Developing energy-efficient AI models through techniques such as model quantization and pruning can reduce computational demands. Optimizing data storage and management, along with utilizing renewable energy sources for data centers, can significantly lower AI’s carbon footprint. Implementing green computing practices, such as dynamic power management and hardware–software optimization, can further enhance energy efficiency. Regular monitoring, maintenance, and lifecycle assessments of AI systems can support long-term sustainability while reducing the need for resource-intensive retraining. By adopting these strategies, healthcare institutions can balance the benefits of AI advancements with environmental responsibility, ensuring that AI-driven clinical applications align with broader sustainability goals (62-64).
4.8. Study Limitations
This study acknowledges certain limitations. A key limitation is the small sample sizes in several studies (38-40), which heightens the risk of overfitting. Overfitting occurs when AI models perform exceptionally well on training data but fail to generalize to unseen cases (53, 54). To mitigate this risk, several strategies can be employed. First, data augmentation techniques — such as rotation, flipping, contrast adjustment, and stain normalization — can artificially expand the dataset, improving the model’s ability to generalize to new cases. Second, transfer learning, where AI models are pre-trained on large-scale histopathological datasets before fine-tuning on smaller OKC-specific datasets, can enhance performance and reduce overfitting. Third, cross-validation methods, particularly k-fold cross-validation, should be used to ensure that the model is evaluated on different subsets of data, thereby reducing bias from reliance on a single training set. Fourth, regularization techniques such as dropout, batch normalization, and L2 regularization can help prevent the model from overfitting to features that may not generalize well. Fifth, leveraging synthetic data generation using GANs can supplement real training samples, particularly for rare cases like OKCs. Lastly, multi-center collaborations and federated learning approaches should be pursued to train AI models on more diverse datasets without compromising patient privacy.
Another limitation was that few studies reported data suitable for meta-analysis, including AUC, sensitivity, specificity, and accuracy. Additionally, in the meta-analysis, the weight of each study was determined by its sample size, reflecting the relative contribution of each study's data volume to the pooled estimate. While this approach is straightforward, it does not account for differences in study precision or variability, which may limit the robustness of the pooled results.
Finally, one study had to be excluded due to the lack of access to its full text, which prevented a comprehensive evaluation of its methodology and results.