1. Background
Chronic lymphocytic leukemia (CLL) is a common blood cancer in people aged over 40 (1). Its prevalence in the Western world, particularly across Europe, North America, and Australia, is 40% of all leukemia cases in people over 65. The prevalence of this cancer is reported to be lower in Asia and is rare in India, China, and Japan (2-5). According to the morphological evaluations, the dominant cancer cells in this type of leukemia are small mature B lymphocytes found in the blood, which exist in greater numbers in the bone marrow, lymph nodes, and spleen compared with normal cells. Most tumor cells spread through the lymphoid organs and blood circulation. As the disease progresses, the lymphoid organs and the lymph nodes gradually enlarge. This clinical symptom is significant for determining the stage and grade of the cancer (6). Malignant cells presenting themselves in patients with CLL have certain markers that distinguish them from different types of lymphomas (7). The main cause of this disease is still unknown, but like many other cancers, it is greatly affected by genetic factors like chromosome abnormalities and genetic mutations as well as environmental factors such as radiation, magnetic field, viral factors, and chemicals such as insecticides and toxins (8, 9).
Immunophenotypic markers of CLL consist of B cell populations increase with the markers of CD19, CD5, and CD23; absence of FMC-7 marker; and low expression of CD22, CD79b, and surface immunoglobulin (10). Approximately 99% of tumor cells are in the G0 or the beginning of the G1 phase of the cell cycle; therefore, it is difficult to conduct cytogenetic tests on these cells in the S phase (11).
In addition to clinical markers based on the appropriate classification systems, detection of different molecular markers are of great value for the prognosis of the disease. Molecular markers consist of determining mutation in the variable region of immunoglobulin heavy chain (IgVH), Fluorescent in situ hybridization (FISH), cytogenetic analysis, and measuring ZAP70 and CD38 protein expression (12, 13).
CLL is a heterogeneous disease in which numerous genetic factors are involved. For example, the mean survival rate in groups with chromosomal deletion (which deletions) ranges between 32 and 133 months (14). The diagnosis and study of these abnormalities is conducted through Array Comparative Genomic Hybridization (aCGH) of the normal and cancer cells as well as through FISH study (15).
In 1999, Hamblin et al. (16) and Damle et al. (17) demonstrated that patients with CLL can be divided into 2 subgroups based on IgVH mutation of BB-cell receptor (BCR) of leukemic cells (18). In fact, CLL is called a naive B lymphocyte (a mature lymphocyte which has not yet been exposed to an antigen in lymph nodes). During the pathogenicity process, the naive CD5+ B cells might undergo somatic mutations in IgVH upon entering the germinal center of lymph nodes (18, 19). Meanwhile, another group of patients have a population of intact B lymphocytes tumor cells that have not entered the germinal center of the lymph nodes (and are still in the pre-germinal center stage), cells that have not been mutated yet and are thus called IgVH unmutated cells (19). The findings of the studies show that prognosis is poorer for patients whose cancer cells have not been somatically mutated.
2. Objectives
This study aimed to determine the frequency of heavy chain mutation in patients with chronic lymphocytic leukemia and examine its relationship with clinical reports and flow cytometric analysis. Through sampling and collecting genomic DNA using the primers defined for IgVH mutation, we detected the genomic DNA mutation by PCR and gene sequencing.
3. Patients and Methods
The study population comprised all CLL patients referred to the central clinic and Valiasr center of Imam Khomeini hospital in Tehran from 2009 to 2013. All patients with CLL disease were included and exclusion criteria were having allergic or other inflammatory diseases. The study was approved by local ethics committee.
Upon registering at the hospital and undergoing clinical examinations, those CLL patients who had no record of chemotherapy signed informed written consents for CBC test (peripheral blood cell count) and participation in the research. Next, their blood samples were collected using 2 heparin tubes marked by each patient’s confidential code.
3.1. Disease Staging Based on Rai and Binet Systems
The patients were followed up by means of further referrals and periodic visits to the hospital or clinic every 3 or 6 months for at least 3 years after the beginning of the research or their demise. CBC and lactate dehydrogenase (LDH) tests were carried out and analyzed during each examination session.
3.1.1. Flow Cytometry Analysis
Upon lysing the peripheral red blood cells, specific monoclonal antibody conjugated with different fluorescent dyes were used against CD23, CD3, and CD10 markers and a dual dye to distinguish CD19 and CD5 markers as well as control antibodies (Dako, Denmark) (1).
3.1.2. Isolation of Peripheral Mononuclear Blood Cells
Patients PBMC were isolated from 5 mL of the peripheral blood by Ficoll (Lymphodex, Inno-Train, Kronberg, Germany) (1).
3.1.3. Chemotherapy Protocols
Some patients in this study have received anti-cancer chemotherapy protocols such as FC: fludarabine and cyclophosphamide; FCR: fludarabine, cyclophosphamide, and rituximab; and FR: fludarabine and rituximab (1). Table 1 shows the percentages of these patients.
| Chemotherapy Protocols | No Drug | FC | FCR | Chlorambucil | FR |
|---|---|---|---|---|---|
| Patients percentage | 60 | 8 | 11 | 6 | 15 |
Abbreviations: FC: fludarabine, cyclophosphamide; FCR: fludarabine, cyclophosphamide, rituximab; FR: fludarabine, rituximab.
aValues are expressed as percentage.
3.1.4. Freezing PBMC
Ficoll separated PBMCs stained by Trypan blue (SIGMA®, Taufkirchen, Germany) and counted by Neubauer slide and resuspended in the culture medium used for freezing viable cells, which is RPMI-1640 (GIBCO, Life® technology, USA), FBS 10% (GIBCO, Life® technology, USA), and DMSO 10% (SIGMA®, Taufkirchen, Germany). The mononuclear cells were stored at -70°C freezer until all samples were successfully collected (1).
3.1.5. Genomic DNA Extraction
Upon transferring the cells from -70°C freezer to 37°C water bath, they were washed with PBS, and upon centrifuging, DNA was extracted on a plate using Genet Bio Kit (Genet bio, Korea). Concentration of the obtained DNAs was measured using a NanoDrop Thermo (Thermo, Thermo Fisher Scientific, and USA) (1, 20).
3.1.6. IgVH Mutational Analysis
For measuring the IgVH mutations, polymerase chain reaction (PCR) was performed by Taq DNA polymerase enzyme using the standard primers defined for IgVH gene families (21). The yielded PCR product was run on electrophoresis agar gel and those with a positive band were isolated with a scalpel blade and their DNA content was isolated from the gel using a Qiagen Kit (QIAGEN, USA). The PCR products were ligated into the pGEM®-T vector (Promega, Wisconsin, USA) and cloned in competent bacteria produced by JM109 high efficiency competent cells through fusion and freezing by T4 DNA Ligase. Next, bacterial suspensions were grown on X-gal medium and white colonies were selected and passaged in an LB culture medium containing antibiotics. Finally, the plasmids of the intended colonies were extracted by Plasmid Midi Kit 100 (QIAGEN, USA) and the specific plasmids were sequenced by ABI PRISM 7700 sequence detection system (Applied Biosystems, USA). After the sequences were determined, the gene bank was searched for the acquired sequence (BLAST) and its mutation rate was studied against somatic cells and germ line normal sequences (1)
3.2. List of Primers Used in Heavy Chain Mutation Analysis
3.2.1. Statistical Analysis
Data analysis was done using SPSS 20 (IBM, SPSS, statistics software). Demographic results were compared and studied using the Chi-square test and the survival data was estimated by Kaplan Meier estimator and Log-Rank test.
4. Results
4.1. Clinical Results
The initial sample included 30 patients. Four of them were excluded due to failure to revisit, their personal request, or improper care of the initial sample. Data of the remaining 26 patients were analyzed. There were 7 female and 19 male patients with CLL, with the mean age of 62 (standard error of 1.84) years and the median age of 64 (standard deviation of 9.14) years. The patients were not under any medications up until sampling started, but 10 of them later underwent chemotherapy during the follow-up period. Table 2 shows the medications they received.
| Staging | Stage 1 | Stage 2 | Stage 3 | Stage 4 |
|---|---|---|---|---|
| Rai | 40.7 | 29.6 | 14.8 | 11.11 |
| Binet | 62.9 | 18.5 | 14.8 | - |
aValues are expressed as percentage.
4.2. Flow Cytometry Results
According to the cell surface marker screening panel, the CD markers, including CD5, CD10, CD23, CD3, and CD19 were analyzed in the patients’ blood samples by flow cytometry to verify their chronic lymphoid leukemia. In this regard, the conformity of the patients to the flow cytometry panel and the mean percentage of the markers were assessed (Table 3).
| CD Markers | CD19 | CD23 | CD5 | CD3 | CD10 | CD5/CD19 |
|---|---|---|---|---|---|---|
| Percentage of patients | 76.12 ± 23.172 | 59.48 ± 26.587 | 51 ± 28.479 | 10.72 ± 11.942 | 0.2 ± 0.5 | 39.08 ± 26.587 |
4.3. IgVH Mutation Analysis Results
A sample of 20 DNAs from 26 patients was sufficient and had good quality for the analysis of IgVH gene mutations. Of 20 samples that were referred to the immunology laboratory of Semnan University of Medical Sciences, 7 received a negative result in the amplification, replication, or the PCR test, and 6 of them were in a heavy chain unmutated status. It was then determined that the most prevalent heavy chain gene pertained to, in descending order, IgVH3 (66.66%), IgVH4 (16.1%), and IgVH1 (11.11%) alleles, but the least occurrence percentage was related to IgVH2 gene. Then, the patients were assessed concerning their Rai stage and their IgVH mutation status, and it was found that out of 7 cases in an IgVH unmutated status, 3 were in stage 1, 2 in stage 2, and 1 in stage 4 of the disease. Table 4 shows the comparison between the patients’ IgVH genes.
| Total | IgVH1 | IgVH2 | IgVH3 | IgVH4 | |
|---|---|---|---|---|---|
| Mutated | 38.90 | 0.00 | 5.00 | 46.00 | 11.11 |
| Unmutated | 61.10 | 11.00 | 0.00 | 22.00 | 5.00 |
Abbreviation: IgVH, immunoglobulin variable heavy chain.
aValues are expressed as percentage.
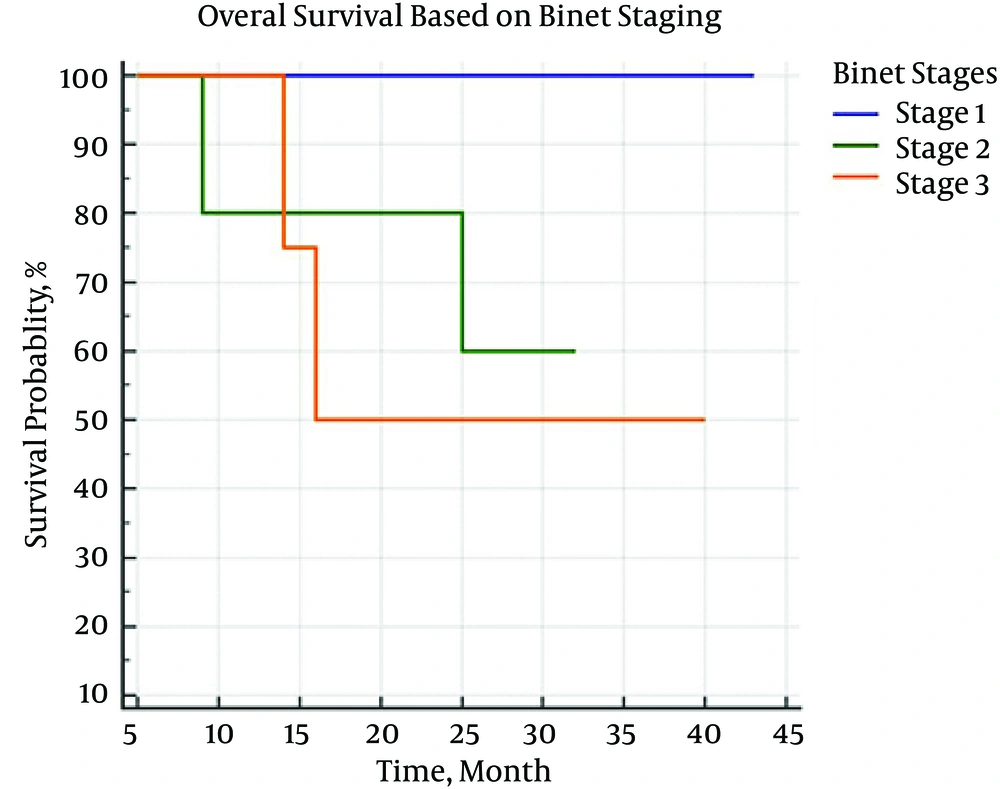
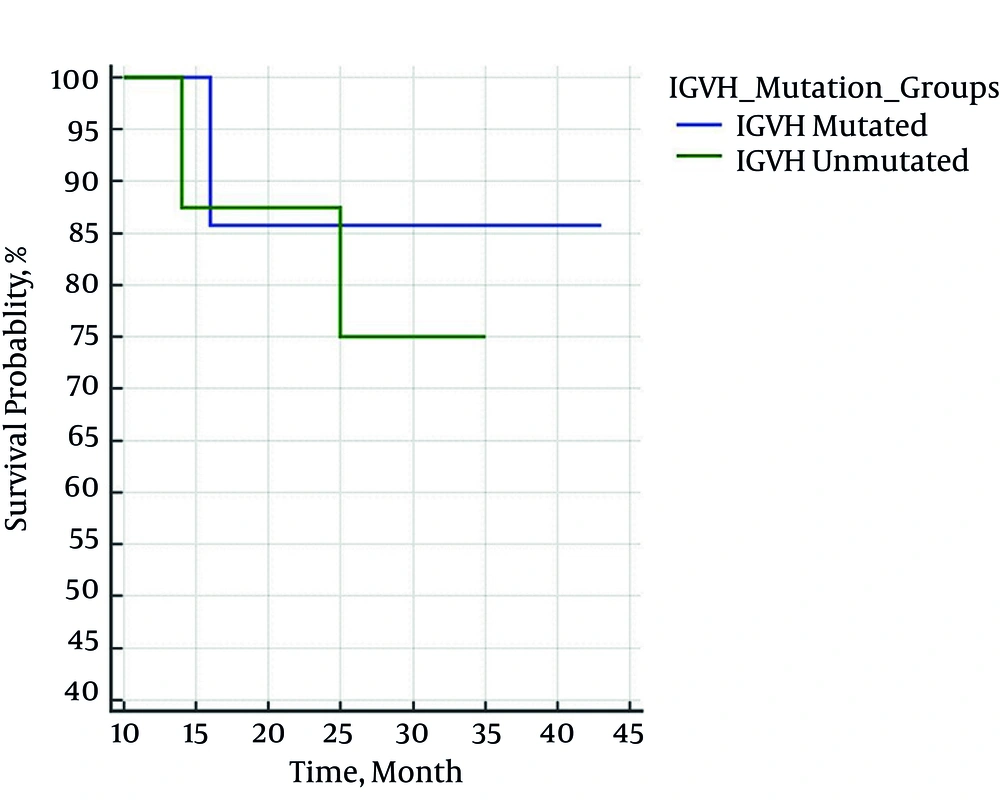
On the follow-up period, 4 patients died. Using Kaplan Meier estimator and Log-Rank test, the mean survival rate of patients based on the presence or absence of IgVH gene mutation was estimated as 39 (32 - 46) and 31 (26 - 36) months, respectively (P = 0.407). The correlation between survival rate and IgVH gene mutation and Binet staging are shown in Figures 1 and 2.
5. Discussion
According to the results of the IgVH mutation analysis, 66.66% of the patients had mutations in IgVH3 alleles with different heterogeneities. Based on a similar study conducted in 1998 on Burkitt lymphoma in Iran, the highest percentage of IgVH mutations in these patients occurred in IgVH3. In the study conducted by Chapman et al. in northwestern Iran, upon accomplishing clinical studies and the other tests, including IgVH analysis, mutation pertaining to the special sequences of Burkitt lymphoma was also studied as a complementary test. By comparing the Chapman et al. study and the present research, we can plan a phylogenic study based on the type of IgVH involved in malignancies (22). In the study conducted in 2007 on patients with CLL, the IgVH of 59 patients were analyzed and the patients’ IgVH mutation percentages were reported to be 45.8% (VH3), 18.6% (VH1), 32.2% (VH4), 1.7% (VH6), and 1.7% (VH5). Similar to the findings of other studies discussed, the highest mutation percentage in this study related to IgVH3 (23). According to the results of the study conducted in 2009 on 87 patients with CLL, IgVH mutations pertaining to IgVH3 were more prevalent (56.4%) while the mutation percentages pertaining to the other types of IgVH were 20.7% (IgVH4), 18.4% (IgVH1), 3.4% (IgVH6), and 1.1% (IgVH5). Similar to other studies, the highest percentage of the IgVH unmutated samples in this study (17%) pertained to patients with IgVH3 mutation.
The prognosis of patients with IgVH3 mutation is reported to be at a critical IgVH unmutated status (21). According to the study conducted in 1993, IgVH3 mutation frequency is also related to factors causing multiple myeloma (24). In a study conducted in 2000 by Gharagozloo et al. on patients with multiple myeloma in Iran, a significant relationship was found between IgVH3 mutations and disease severity. The frequency of B cell subtypes was also examined concerning the IgVH mutation type, and it was found that IgVH3 is significant in this case (25). Therefore, based on the findings of this study and similar studies in Iran, there is a direct relationship between IgVH3 and disease severity, and the percentage of patients who have a poor prognosis due to being IgVH unmutated is higher in this group. Because of the high cost of IgVH mutation analysis, the sampling was performed in a small number of patients, which is a major limitation for statistical analysis.
In both the present and other similar studies, mutation is more prevalent in IgVH3. Although the presence or absence of mutation affects the patients’ survival rate, this effect might be statistically insignificant because of the small sample size. Nevertheless, there was a significant relationship between survival rate and the clinical stage of the disease in the present study. We can conclude that clinical methods are still valuable in determining the prognosis of patients with CLL. Given the high costs and the need for special laboratory, it is necessary to determine the cost-effectiveness and value of examining IgVH mutations and determining the types of mutation, such as IgVH3, in more extensive studies.
Limitation of this study was small sample size due to the lack of adequate budget.
.jpg)