1. Background
Gene expression investigations in human diseases provide useful diagnostic and prognostic approaches for many disorders (1-3). Tissue specimens can be kept for a long time after fixing in formalin and embedding in paraffin (FFPE). Nowadays, formaldehyde is used as a common chemical tissue fixator, which is widely used due to low cost and minimal changes in cell and tissue structure (4, 5). To preserve the tissue structure during fixation in formalin, the tissues are embedded in paraffin. Currently, RNA extraction from FFPE tissue specimen is one of the fundamental challenges in gene expression profile studies (6). There is no gold standard as the most efficient method for RNA extraction from FFPE specimens. Ten percent formalin solution is routinely used to fix tissues in pathology labs. Formaldehyde primarily reacts with the ε-amine-N group of lysine protein residues, resulting in the formation of amino ethanol moieties, which in turn reacts less with amide- and guanidyl-N groups. It modifies ring structures of Tyr, Trp, and His residues as well (7). The reaction of formaldehyde with Nucleic Acids (NA) is known to involve exocyclic amino groups of A, C, and G residues (5, 8), due to the effect of formaldehyde on adenine and other nucleic acid bases that methyl bridge produces. This avoids complementary base pairing during analysis of molecular hybridization reactions. The histological cross-linking between the nucleic acids and other macromolecules reduces the efficiency of RNA extraction (9). The aim of this study was to evaluate factors, which can effect RNA extraction yield from FFPE samples using High Pure Paraffin Kit from Roche Company (10) to improve the RNA extraction protocol.
This study examined the following objectives: 1) Effect of specimen’s storage time on RNA recovery from FFPE samples; 2) Effect of tissue section size on RNA recovery from FFPE specimens; and 3) Effect of proteinase K concentration on extracted RNA quality from FFPE specimen.
2. Material and Methods
After Semnan University of Medical Sciences ethical committee approval (ethic code: 598), tissue specimens were obtained from 78 patient biopsies, which were stored at the Pathology Lab (Kosar Hospital, Semnan, IRAN) archive. Informed consent was given by all participants in keeping with the Helsinki declaration (11). Pathologic specimens from breast cancer patients and normal tissue were classified by a pathologist.
2.1. RNA Extraction
RNA extraction from FFPE was performed by a kit from Roche Company (High Pure RNA Paraffin Kit # Lot: 03270289001, Basel, Switzerland). Paraffin removal and tissue slicing were performed in accordance with the proposed kit protocol: FFPE specimens were sectioned in 1x10 and 5x2 micrometer by microtome, collected in RNase free tubes, and 800 micro liters of xylene was added to each tube to remove the paraffin. In total, 400 micro liters of absolute ethanol was added and centrifuged with 12 to 14×1000g the sediment, washed with 1 mL of absolute ethanol, and incubated overnight for tissue breakdown (recommended kit protocol) (10). Finally, RNA was eluted from RNA extraction column by adding 50 mL of DEPC water.
2.2. Reverse Transcription (RT) Reaction
According to kit recommendations, 10 µL of total RNA, which was extracted from FFPE specimens, was reverse transcribed in a final volume of 20 μL, using all components from the RT kit (Revert Aid First Strand cDNA Synthesis Kit #K1622 Lot:00195472, Thermo Fisher Scientific, Waltham, Massachusetts, USA). Extracted RNA was added to 1 μL of oligo (dT) primer and 1 μL random hexamer primer; final volume was adjusted to 12 μL with DEPC water, the incubation time was for 5 minutes at 65 °C. After incubation, 4 μL of RT buffer, 1 μL RiboLock RNase Inhibitor (20 u/μL), and 2 μL 10 mM dNTP Mix and RevertAid H Minus M-MuLV Reverse Transcriptase (200 u/μL) was added and incubated for 60 minutes at 42°C. Afterwards, the reaction was terminated by heating at 70°C for 5 minutes. All RT reactions were done in a PCR thermocycler (Eppendorf, Hamburg, Germany) for 5 minutes at 65°C, 60 minutes at 42°C, and 5 minutes at 70°C following the manufacturer's recommendations. Finally, the cDNA was stored at -20°C.
2.3. Primers
Primers were designed with exon junction so that the product length was approximately 161 bp and melting temperature was adjusted between 60 and 63°C, and GC count was between 40% and 70%. Primers were designed with primer 3 web software and primers were blasted with Humosapian gene sequences from National Center for Biotechnology Information (NCBI) GenBank database (www.ncbi.nlm.nih.govweb site). Species homologies were checked in gene sequences from Ensemble GenBank (http://www.ensembl.org/index). Primer-dimers and primer secondary structures were checked with the OligoCalculator web software (http://www.basic.northwestern.edu/biotools/oligocalc). Primer sequences are listed in Table 1.
| Primers | Sequence | Length |
|---|---|---|
| β-actin sense | CGACAGGATGCAGAAGGAGA | 20 bp |
| β-actin antisense | CGTCATACTCCTGCTTGCTG | 20 bp |
| Product length | 161 bp |
β-Actin Primers Sequence
2.4. Quantitative Real Time Polymerase Chain Reaction Assays
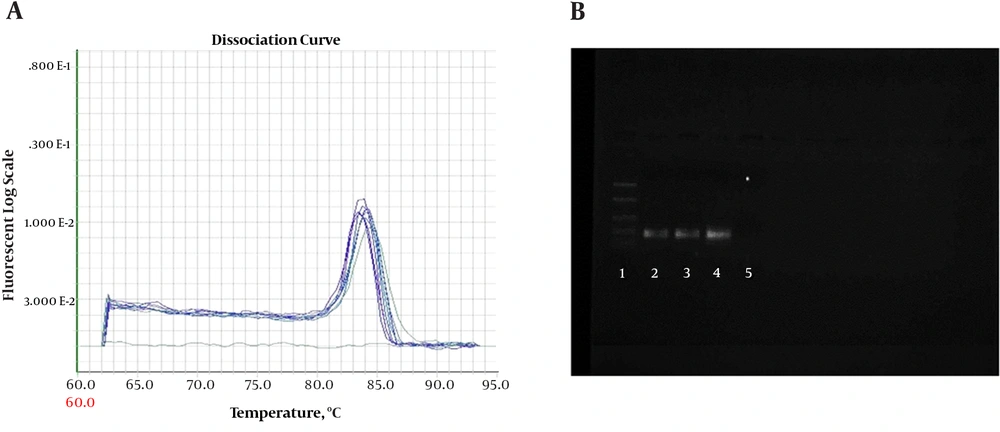
Quantitative PCR (QPCR) reactions were conducted on ABI 7900 (7900HT Real-Time PCR System with 96-Well Block Module, USA). Amplifications were performed with SYBR Green QPCR Master kit (MicroAmplicon master green high rox-Denmark-ID-No-5000840-1250). Reactions were adjusted in a final volume of 20 μL with 1μL of cDNA and 0.5 μL of each primer 10 pM (Sense and Anti Sense) concentration. Amplifications were performed for conventional PCR assays. A melting curve was included for all reactions to confirm Tm of each amplicon. To check the formation of primer-dimers or contamination, NTC (no-template control) was included in each assay. The PCR reactions were performed on ABI 7900 Thermal Cycler. The PCR thermal profile consisted of 2 minutes at 50°C, 10 minutes at 95°C, followed by 50 cycles of (30 seconds at 94°C and 1 minute at 60°C). The PCR products were checked on 1.5% agarose gel (Figure 1).
A, Melting curve for β-actin PCR products. Curves showing positive real-time PCR amplification of the β-actin gene; B, Gel electrophoresis for PCR products. Amplification was detectable in the samples run in lanes 2 to 4, with product corresponding to a 161-bp band, Lane 1 was the 100-bp ladder, and lane 5 contained the non-template control with DNase/RNase-free water.
2.5. SYBER Green PCR Reaction Efficiency
A serial dilution (10-fold) in 8 points was performed for determining SYBER Green quantitative PCR reaction efficiency. Calibration curves were plotted for calculating efficiencies and correlation coefficients.
2.6. Effect of Specimen Storage Time on RNA Recovery
To investigate the effect of storage time on RNA extraction yield from formalin fix paraffin embedded tissues, quantity and quality of total RNA was compared in samples with a lifespan of 1 to 3 years, and a few days to 12 months. Therefore, after cDNA preparation in all specimens with the same condition, β-actin gene mRNA expression in all samples (with the same amount of RNA-1000 ng) was compared in Real Time PCR by specific primers (Table 1) and Ct (cycle threshold) value.
2.7. Optimization of Tissue Section Size
Different tissue section sizes may influence RNA extraction yield. Tissue sections with 10-micrometer thickness versus 5 sections with 2-micrometer thickness were used for RNA extraction (n = 10 samples were randomly selected and RNA were extracted by 2 section size methods). Furthermore, 10 μL of total RNA (extracted by these 2 methods) was converted to cDNA in the same condition and B-actin Ct was used to evaluate RNA.
2.8. Proteinase K Effect on RNA Extraction
To investigate the effect of proteinase K on RNA extraction yield, proteinase K from 2 different companies was purchased and RNA was extracted with proteinase K from Bioneer (8-11 Munpyeongseo-ro Daedeok-gu Korea, Republic of (South)) and Roche (Basel, Switzerland) Companies, alone or together. RNA extraction without any proteinase K, was also examined.
2.9. Gel Electrophoresis
The RNA extract was visualized by 1.5% agarose gel electrophoresis and stained with ethidium bromide and the results were analyzed by the Uvtec instrument (House 36a- Cambridge, UK), Fire Reader XS, software (France).
2.10. Data Analysis
Data were analyzed and processed with the SPSS 16 software on a Windows operating system. Gene expression level was compared using the Mann-Whitney U-test. A P value of less than 0.05 was considered significant.
3. Results
3.1. The Effect of Storage Time on RNA Recovery
Comparing Ct value of β-actin genes in tissue samples with different lifespan indicated that old samples had higher Ct values, which indicates less initial copy number of β-actin mRNA. Using regression statistical analysis between age and Ct value, a positive correlation (0.38) between time and Ct values of β-actin RNA was obtained meaning that RNA extraction yields will decrease with increasing storage time of tissue samples.
3.2. Optimization of Tissue Section Size
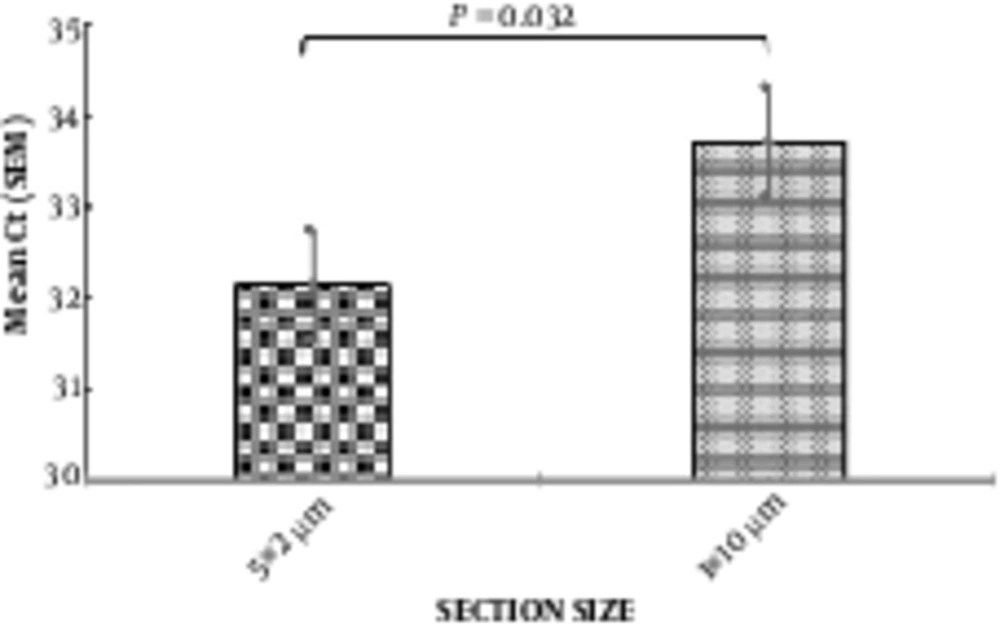
Comparing the amplification of β-actin gene, using real-time PCR, for 2 methods of RNA extraction (1 × 10 micron slices and 5 × 2 microns), indicated that 2 sections with small thickness of 5 µm versus one section with 10-µm thickness had better results for RNA analysis from this tissues, and increased the yield of RNA (Figure 2).
Comparison of the effect of two methods for tissue hemogenetion (2 µm vs 10 µm section size) on RNA extraction yield using β-actin gene expression (n = 10). In the left column the mean CT for thin sections (2 µm) was 31 and in the right column the mean CT for thick sections (10 µm) was 33.71. CT: cycle Threshold, N: samples
3.3. Proteinase K Effect on RNA Extraction
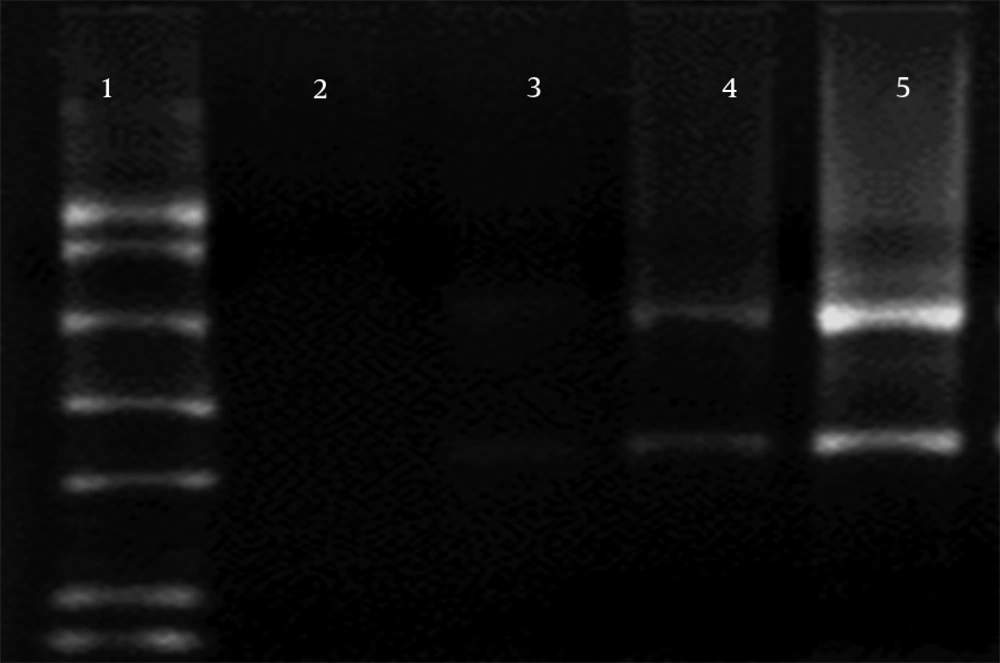
Three separate experiments, 1) Proteinase K from Roche alone, 2) together with Proteinase K from Bioneer, and 3) no Proteinase K, were used for investigation of the effect of Proteinase K on RNA degradation during RNA extraction, and the results were compared (Figure 3). In electrophoresis samples with no Proteinase K, RNA was not found in 1.5% agarose gel (lane 2). However, preparation with Proteinase K (20 mg/mL) from Bioneer and Roche was used to show 28S RNA band and smear in the gel (lanes 2 and 3). Using both Proteinase K from Roche (10 mg/mL) and Bioneer (10 mg/mL) together showed stronger 28S and 18S RNA band in agarose gel electrophoresis (lane 5).
4. Discussion
Formalin-fixed paraffin embedded tissues are used for pathologic diagnosis and are archived for a long period of time. These samples could be used for molecular research and diagnostic purposes. However, this type of preservation makes it difficult to perform molecular analysis on these type of samples due to nucleic acid modification by significant protein-protein and protein-nucleic acid crosslinks, following formalin/paraformaldehyde fixation. The fixation process itself doesn’t cause nucleic acid fragmentation (12, 13), instead, the embedding process, which requires high temperature and vacuum for the paraffin penetrance to the tissue, may lead to chemical reactions, which subsequently modify RNA and DNA. These modifications can cause fragmentation over time (14).
Investigation of gene expression profile can be performed by detecting specific RNA expression levels, using a variety of methods, including microarray analysis, qPCR, and southern blotting. mRNA comprises only of 1% to 3% of total RNA, by a weak extracting method from FFPE samples, gene expression pattern is not readily detectable in this sample even with the most sensitive methods. On the other hand, ribosomal RNA makes up > 80% of total RNA samples, with the majority comprised by the 28S and 18S rRNA species (in mammalian cells).
In this study, the researchers sought to obtain a suitable protocol for RNA extraction FFPE tissue by using RNA extraction kits from the Roche Company. The study compared the RNA extraction method proposed by Roche kit with current conventional methods, such as tissue slices, as well as the proteinase K.
Mark Abramowitz et al. in their study, indicated that proteinase K could improve RNA extraction yield (15). In this study, the researchers attempted to use a combination of 2 different types of proteinase K to investigate the effect of Proteinase K on RNA degradation during the RNA extraction process. Comparing Intact RNA electrophoresis on agarose gel indicated that 28S and 18S rRNA bands resolution was increased by adding proteinase K (eukaryotic ribosomal RNA).
Commonly, 28S rRNA band should be about twice as strong as the 18S rRNA band. This 2:1 ratio (28S:18S) shows that the RNA is completely intact. RNA from FFPE samples is usually more degraded than RNA from fresh tissues. Partially degraded RNA appears as smeared or does not exhibit the 2:1 ratio of high quality RNA (16). The current results showed that combination of 2 proteinase K increases the yield and also reduces the degradation of RNA. Proteinase K can improve the efficiency of RNA extraction by rapid degradation of DNase and RNase released from the tissue during the RNA extraction process (17).
This study also checked the effect of tissue storage time on RNA extraction yield. In previous studies scientists showed that formaldehyde crosslinks with amino groups in nucleic acid bases and proteins in irreversible manner does not degrade RNA (18). However, over time, the methylene bridges of formaldehyde with proteins and DNA will increase. This does not effect the length of the extracted RNA yet it has a negative effect on the performance of reverse transcription enzyme, by disrupting base pairing and the process of converting RNA to cDNA. To clarify this effect, this study considered the β-actin gene amplification in 78 formalin fixed tissue samples with a lifetime of 1 to 3 years and less than 12 months. B-actin mRNA quantification indicated that samples with less than 12 months had a better quantity of mRNA than samples with 1 to 3 years of lifespan. In a similar study in 2014, Soo Kyung et al. reported that long storage time limits the RNA extraction yield (19). However, Kokkat et al. in their study in 2013 reported that long storage time had no effect on RNA extraction yield from lung, salivary glands, and thyroid tissues (20). The following recommendation should be considered if one wants to use FFPE specimens for gene expression analysis, in addition to histopathology: RNA should be isolated within 1 year after embedding, i.e. before degradation during storage of FFPE samples and the application of this sample should be limited (18).
Some kits use column filters to extract RNA. Enormous amounts of tissue can close the pores of these filters, resulting failure to extract RNA. Also, to extract RNA from tissue, it should be thoroughly homogenized to increase the efficiency of RNA extraction and prevent clogging of filters (21). The remarkable finding of this study was that less time of incubation for extracting RNA led to less tissue breakdown. By applying this effect, the efficiency of RNA extraction will increase.
In this study, the researchers tried to keep a constant volume of tissue mass in accordance with the kit recommendations (10). It was attempted to provide more homogeneous tissue with smaller sections. The results indicated that a smaller section size could improve extraction efficiency (Figure 2). According to this study, homogeneity of paraffin samples as well as the use of proteinase K could increase the RNA extraction efficiency. In this study, FFPE was used from breast tissue specimens and it was assumed that similar results could be obtained from other tissue specimens.
.jpg)