1. Background
Human PTHrP is the product of a single gene and is transcribed into different isoforms through an alternative splicing process that confers its pleiotropic activity (1). The N-terminal domain of PTHrP protein is very similar to that of parathyroid hormone (PTH) and acts on the same receptor associated with G proteins (2). Mature PTH is composed of 84 amino acids, is synthesized by parathyroid glands, and acts on kidney and bone to regulate calcium homeostasis in the blood.
The first evidence of PTHrP protein was observed in cancers (3) associated with increased serum calcium levels and its coding mRNA was isolated from cancer cell lines (4). PTHrP is a paracrine/autocrine factor with more diverse functions compared with those of PTH, which has fewer targets in bone and kidney. PTHrP is required to activate growth of mammary glands, tooth eruption, homeostasis of hair follicle development and is considered as a key factor for the normal development of osteochondral growth plate as well as alleviation of hypertrophy.
Hypertrophy as a natural phenomenon during terminal differentiation of mature chondrocytes, results in the expression of hypertrophic markers such as type X collagen (ColX) and alkaline phosphatase (5), and leads to an increase in cell size. The recent studies confirmed the positive effect of PTHrP on chondrogenic differentiation of marrow/adipose derived mesenchymal stem cells (MSCs) (6). Moreover, some anti-hypertrophic effects are reported for 1-34 N-terminal amino acid sequence of PTHrP (7).
MCF-7 is a human cancer cell line isolated from breast cancer and has some similar features with mammary glands epithelial cells, such as sensitivity to estrogen (8) and PTHrP (9). In addition, PTH/PTHrP cell surface receptors are responsible to induce cell proliferation (10) of breast cancer cells and incidence of bone metastasis (11).
Despite the important roles of PTHrP in breast cancer malignancy as well as chondrogenic differentiation of MSCs, there is no commercial or acceptable procedure to analyze biological activity of its recombinant form. Given the high sensitivity of cell line MCF-7 to PTH/PTHrP, which results in proliferation in response to the sub-nanomolar concentration of the protein, it has good potential to investigate PTH/PTHrP biological activity.
2. Objectives
In the current study, the PTHrP encoding gene was isolated from a cDNA library of the human testis, cloned and expressed in Escherichia coli host. The biological activity of the recombinant protein was investigated using a sensitive and reproducible method, which assayed PTHrP-mediated cell line MCF-7 proliferation. To prove the efficiency of the new method, anti-hypertrophic properties mediated by PTHrP during chondrogenic differentiation of MSC was tested. The efficient procedure of bioassay used to analyze recombinant PTHrP could also be utilized as a practical and robust method for biological evaluation of this protein.
3. Methods
3.1. Gene Isolation and Cloning
The coding sequence of mature PTHrP protein “aa 37-177” (UniProtKB: P12272) was amplified using a cDNA library isolated from human testis with primers designed to have NCOI and XhoI at N-terminal and C-terminal ends, respectively (Table 1). The amplified sequence was cloned in pET32a (+) vector (Novagen Inc., Madison, Wis) using digestion/ligation cloning method and transformed into competent cells of E. coli DH5α. The success of cloning was verified using T7 colony polymerase chain reaction (PCR) amplification and DNA sequencing. Next, the plasmid containing correct insert was extracted and transformed to Rosetta™ 2(DE3) pLysS competent cells (Novagen 71403).
| Gene | Forward Primer (5’ - 3’) | Reverse Primer (5’ - 3’) | Amplicon, bp |
|---|---|---|---|
| PTHrP | ACAACACCATGGGCGCTGTGTCTGAACATCAGCTC | ACGATACTCGAGTCAATGCCTCCGTGAATC | 452 |
| ColX | AGTTCTTCATTCCCTATGCCA | CAATGTCTCCTTTCGGTCCA | 267 |
| ALAP | GCCGCAAGTATATGTATCCCA | GCTCAAAGAGACCCAAGAGG | 210 |
| GAPDH | GACCACCATCCATTCCTACAC | GCAAGTCAGGTCCACAACAG | 217 |
Primer Sequences Used for Gene Isolation and qPCR
3.2. Purification and Characterization of Recombinant Protein
The expression host was grown in LB culture media (400 mL) containing 15 µg/mL ampicillin until OD600 = 0.7, followed by induction by 0.2 mM IPTG for 6 hours at 37°C. Using centrifugation, the cells were harvested (1.3 g wet weight), resuspended in 16 mL cell lytic buffer (20 mM PBS, 500 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 100 µg/mL lysozyme, benzonase 80 U/mL, and 0.1% Triton X-100), and left for 1 hour at room temperature. The lysed cells were sonicated followed by centrifugation at 14000 g for 30 minutes. The supernatant containing His-tagged recombinant proteins were purified using Ni-NTA agarose column (Qiagen 30230) washed using wash buffers (300 mM NaCl, 50 mM NaH2PO4/Na2 HPO4 pH 8.0, and 20 or 50 mM imidazole) and eluted using elution buffer (300 mM NaCl, 50 mM NaH2PO4/Na2 HPO4 pH 8.0, 300 mM imidazole), according to manufacturers’ instructions at native conditions. The purified proteins were analyzed on a 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and isolated from stained gel for further mass spectrometry analysis.
3.3. Biological Activity Analysis
The biological activity of PTHrP was measured by its ability to promote the proliferation of human MCF-7 cell line (9). The MCF-7 cell line was obtained from the Iranian biological center (IRBC code: C10062). The cells were cultured in Dulbecco’s modified eagle’s medium (DMEM-F12) (Invitrogen, 21331-020) supplemented with 10% fetal bovine serum (FBS; Gibco Inc. Canada) and 50 mg/mL non-essential amino acid (NEAA) (Gibco Inc. Canada). When cells reached 80% confluence, they were trypsinized and plated in a 96-well culture plate.
Serial dilutions of recombinant PTHrP (0 - 1000 nM) were provided in a 96-well flat-bottom plate in assay medium, then 2000 MCF-7 cells were added to the plate and incubated for 3 days at 37°C in a humidified atmosphere containing 5% CO2. The rates of cell proliferation were assayed using the cell titer 96 non-radioactive cell proliferation MTS (3-(4, 5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; MTS (a) assay kit (Promega). Cell proliferation was assessed by adding 20 µL of the MTS reagent into each well and cells were incubated at 37°C for 3 hours. After this time the absorbance was measured at 490 nm using a microplate reader (Multi spectrum, Themo scientific).
3.4. Chondrogenic Differentiation of MSCs
Chondrogenic differentiation of MSCs was accomplished following a previously reported protocol with minor modifications (12). Briefly, rabbit bone marrow derived MSCs were expanded and trypsinized at passage 3, and then, collected via centrifugation. The cells were resuspended in chondrogenic medium (Lonza, PT-3003) containing 10 ng/mL transforming growth factor (TGF) beta 1 (R and D Systems, 240-B), 200 ng/mL BMP-2 (Sigma, H4791), counted and pelleted in non-adherent 96-well V-shape bottom plate (Corning life science, 3956) through centrifugation at 1000 g for 5 minutes. The plates were incubated at 37°C humidified CO2 incubator for 30 days and the medium was changed every 2 - 3 days. Recombinant PTHrP (0.1 nM) was added to differentiate the medium on day 16 and pellets were harvested on days 23 and 30 for further molecular and histological analysis.
3.5. Histology and Immunohistology Assay
Micro mass pellets were incubated in Boen solution for 6 hours, and then, fixed in phosphate-buffered saline (PBS) containing 2% paraformaldehyde overnight at 4°C. The pellets were dehydrated using tissue processor device (tissue processor DS 2080/H, DID SABZ Co.), embedded in paraffin and cut into 5-µm sections for histological analysis.
Immunohistochemistry analysis was performed on 5-µm section of micro masses after paraffin removal and rehydration. The sections were then incubated with 0.25% trypsin/1 mM EDTA for 5 minutes at 37°C for antigen retrieval. The primary antibody incubation was performed overnight at 4°C and the secondary antibody was applied on sections for 2 hours at room temperature using 1/300 collagen type X antibody (Abcam, ab58632) as the primary and 1/200 goat anti-rabbit IgG (Abcam, ab6721) as the secondary antibody, respectively. DAB substrate was used to detect reaction.
Histochemical analysis of alkaline phosphatase activity was conducted using the qualitative method. Briefly, 10-µm sections of fresh/frozen micro mass were prepared using a freezing microtome device (Leica CM1850) and stained using leukocyte alkaline phosphatase kit (Sigma, 86R), according to the manufacturer’s instruction.
3.6. Quantitative Analysis of Gene Expression
Micro mass RNA was extracted employing RNeasy Mini Kit (Qiagen, 74104) and cDNA synthesis was performed using PrimeScript™ first strand cDNA synthesis kit (TAKARA, 6110A), according to the manufacturer’s instruction. The mRNA levels of alkaline phosphatase (ALAP) and ColX genes were measured by quantitative real time polymerase chain reaction (RT-PCR) (Step one RT PCR Applied BIOsystems, USA) using SYBR® Green PCR Master Mix (invitrogen) as hypertrophic markers. The RT-PCR condition was as follows: incubation at 95°C for 2 minutes followed by 35 cycles at 95°C for 15 seconds and at 60°C for 60 seconds. All data were normalized to an endogenous control (GAPDH) and relative to a calibrator (samples without treatment) and the quantitative results were derived using the 2-∆∆Ct method. The primer sequences are provided in Table 1.
3.7. Statistical Analysis
Statistical analyses were conducted using Prism 5 software. Each experiment was performed in triplicate and analyzed using one-way ANOVA followed by t test. The results were expressed as mean ± standard deviation (SD). In all the cases, a P value of < 0.05 was considered as statistically significant.
4. Results
4.1. Recombinant PTHrP Cloning and Expression
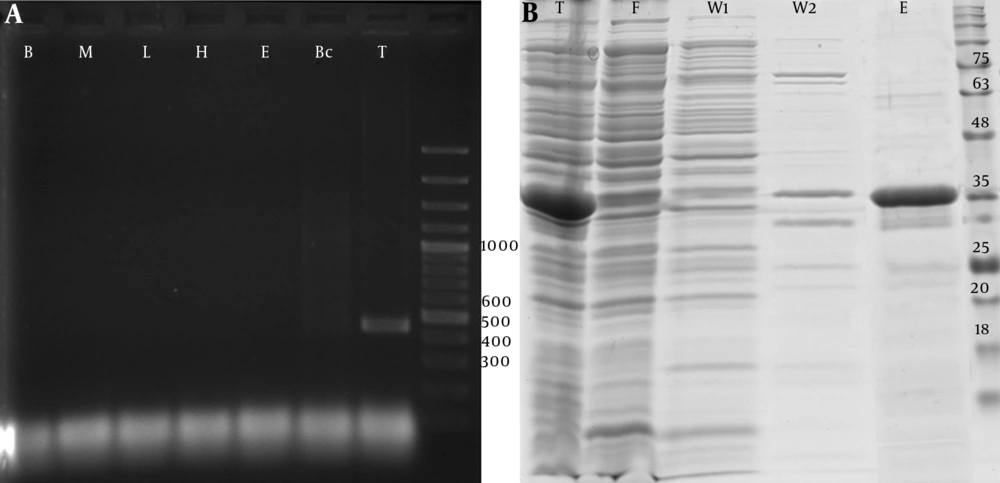
The PCR-amplified fragments of PTHrP with the desired product size that contained the restriction sites for NCOI/XhoI were cloned into pET32a vectors using digestion-ligation cloning method. Colony PCR amplification via T7 primers confirmed the recombinant colonies. Also, DNA sequencing and BLAST analysis of the insert sequence against GenBank verified the sequence of insert and identified it as NM-198965.1 (Figure 1A).
Protein expression analysis showed an increase in the production of recombinant protein in the total cell soluble protein after isopropyl β-D-1-thiogalactopyranoside (IPTG) induction. Protein purification using Ni-NTA column effectively purified recombinant PTHrP protein with high affinity. The summary of protein purification steps is presented in Table 2. Moreover, SDS-PAGE analysis (Figure 1B) of the extracted protein fractions showed a sharp band of approximately 35 kDa excised and identified as PTHrP (UniProtKB-P12272) using MALDI/TOF-TOF mass spectrometry.
A, PCR amplification analysis of the 426 bp fragment of PTHrP coding cDNA derived from human cDNA libraries of blood cells (B), bone marrow derived MSCs (M), liver tissue (L), hepatocytes cells (H), embryonic stem cells (E), breast cancer cells (Bc), and testis tissue (T), b, SDS-PAGE analysis of recombinant PTHrP expressed by pET 32a transformed Rosetta™ 2 competent cells at 37°C for 6 hours. Total soluble cell expressed proteins (T) were purified by 6xHis/Ni-NTA system, washed with 20 (W1) and 50 (W2) mM wash buffers and eluted (E) using 300 mM elution buffer. F: Flow-through fraction of Ni-NTA resin loaded soluble protein.
| Fraction | Total Proteina, mg | Target Proteinb, mg | Purity, %c | Yieldd |
|---|---|---|---|---|
| Insolublee | 28.8 ± 1.1f | 7.3 ± 0.3 | 25.1 ± 0.3 | 1.7 ± 0.0 |
| Soluble | 95.7 ± 4.9 | 28.8 ± 1.5 | 30.0 ± 1.2 | 6.7 ± 0.3 |
| Flow through | 61.0 ± 5.2 | 7.9 ± 0.7 | 12.9 ± 0.1 | 1.8 ± 0.2 |
| Wash 1 | 14.4 ± 1.5 | 1.2 ± 0.1 | 8.5 ± 0.5 | 0.3 ± 0.0 |
| Wash 2 | 1.1 ± 0.2 | 0.3 ± 0.0 | 25.2 ± 2.3 | 0.1 ± 0.0 |
| Elution | 17.0 ± 2.6 | 14.4 ± 2.2 | 95.2 ± 5.3 | 3.8 ± 0.6 |
Purification of rhPTHrP from 1 Liter of E. coli Culture
4.2. Investigation of PTHrP-Induced Proliferation of MCF-7
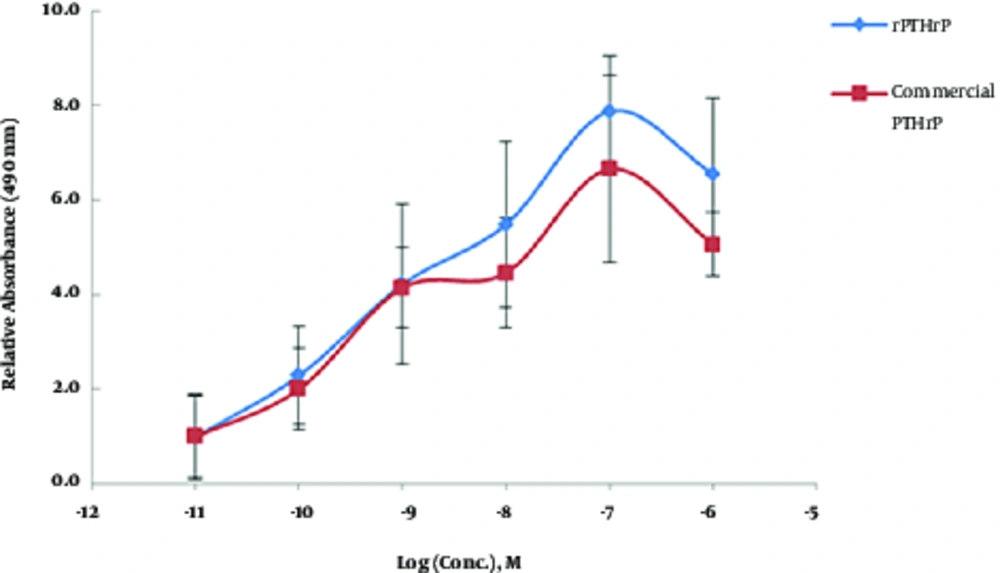
MCF7 is a breast cancer derived cell line and contains cell surface receptors for PTHrP proliferated in response to PTHrP. The current study used MTS analysis (Figure 2) of recombinant PTHrP treated MCF-7 cells to calculate the ratio of cell proliferation in the treated groups to those of the control group (0.01 nM PTHrP treated group). The net OD490 value (∆Rn) was measured by subtracting OD 490 nm of blank (0 nM PTHrP) from each of the PTHrP treated groups. Then, the values were normalized by dividing the subtracted values by the values of the control group and expressed as relative absorbance at OD 490 nm (∆Rn / ∆Rctrl ). The results showed a significant increase in MTS assay absorbance of MCF-7 cells treated with either submicromolar concentrations (1 nM ≤ absorbance ≤ 1 µM) of rPTHrP or commercial PTHrP.
4.3. Analysis of Glycosaminoglycan Content
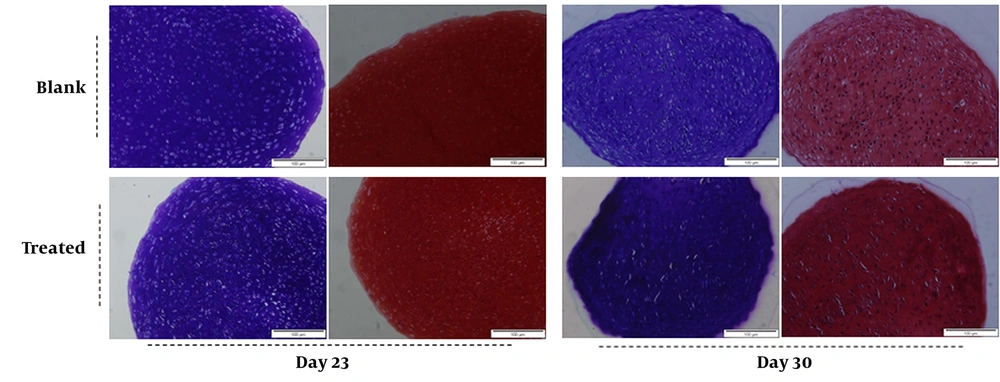
Glycosaminoglycan content of differentiated micro-masses represents chondrogenesis process that can be evaluated using metachromatic staining with toluidine blue and safranin O. Metachromatic staining of s-GAG did not exhibit significant changes in the PTHrP treated group as compared with those of blank either on day 23 or day 30 (Figure 3). Therefore, PTHrP did not show any adverse effects on chondrogenic differentiation potential of MSCs.
Chondrogenic differentiation potential of rabbit bone marrow derived MSCs treated with/without recombinant human PTHrP. The metachromatic staining with toluidine blue and safranin O shows representative analysis of 3 independent experiments. There is no obvious difference between 2 groups (the blank: no PTHrP and the treated: 0.1 nM PTHrP) for metachromatic staining.
4.4. Expression Analysis of Alkaline Phosphatase and Type X Collagen
The activity of ALP and ColX is considered as a specific marker of hypertrophy. Therefore, the expression of ALP and COL10A1 genes were investigated both at mRNA and protein levels using quantitative RT-PCR and histology analysis.
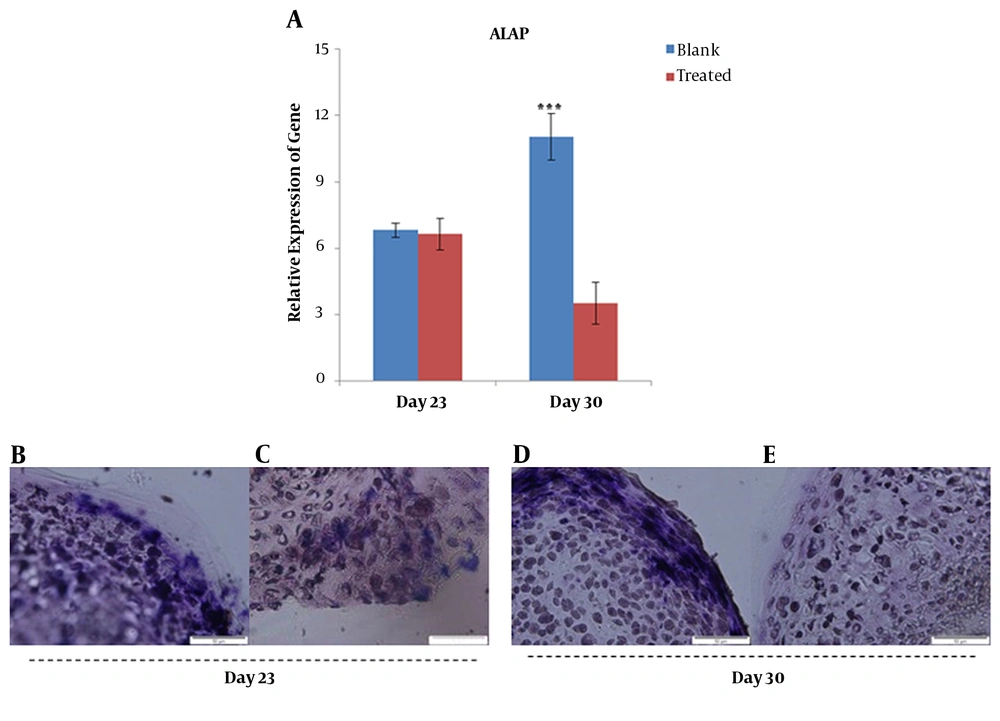
mRNA expression analysis of ALP gene in the blank and treated groups revealed a significant decrease in gene expression in the treated group compared to the blank group on day 30 (Figure 4A). A high intensity of blue color detected in the blank group (0 nM PTHrP) of micro-masses indicated a dramatic activity of alkaline phosphatase of the blank group in comparison with that of the treated group on day 30 (Figure 4B - E).
Gene expression analysis of ALP at mRNA and protein levels; quantitative RT-PCR data for ALP gene obtained after 21 and 30 days for blank and treated groups cultured in chondrogenic differentiation media supplemented with/without PTHrP (A). Histochemistry analysis of 5-µm sections of chondrogenic differentiated micro masses treated with (C and E) and without (B and D) PTHrP. ***; P < 0.001.
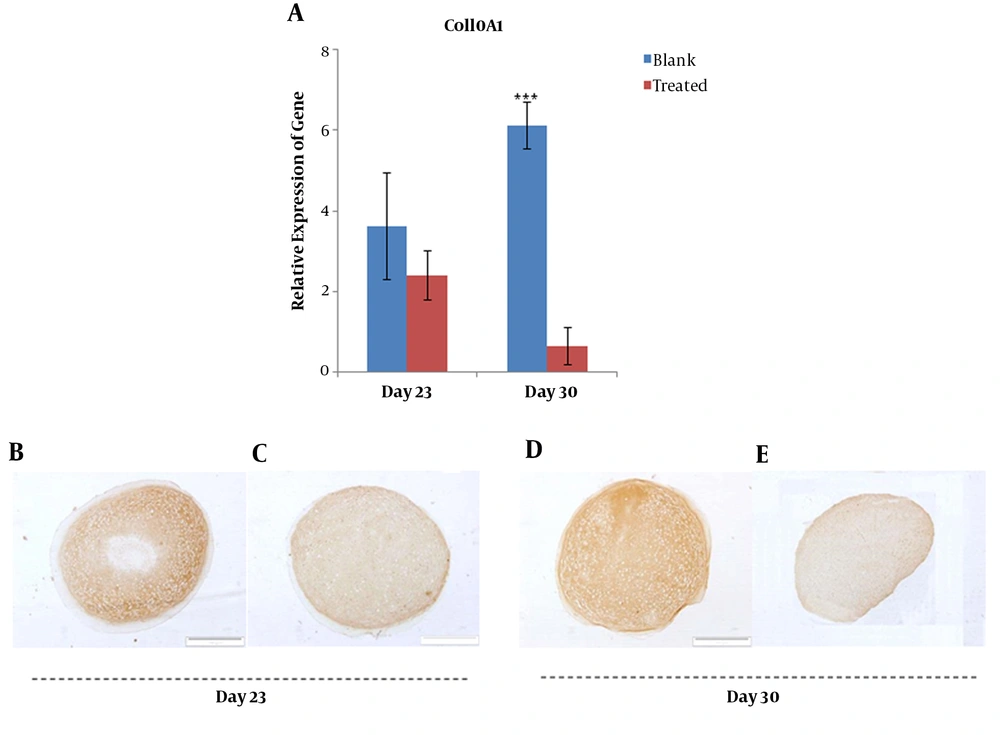
Additionally, mRNA level of ColX exhibited a significant decrease (P < 0.01) in the treated group compared with that of the blank group on day 30 (Figure 5A). At the protein level, ColX did not show a significant change in the expression between days 23 and 30 in the blank group (Figure 5B and D), whereas it drastically reduced in the PTHrP treated group on day 30 (Figure 5C and E).
Gene expression analysis of ColX at mRNA and protein levels; relative gene expression of COL10A1 was quantified using qRT-PCR for blank and PTHrP treated groups at days 23 and 30 (A). Immunohistochemistry analysis of collagen type X protein in micro masses differentiated to chondrocytes with (C and E) and without (B and D) PTHrP at the day 23 and 30.
5. Discussion
PTHrP is an autocrine/paracrine regulatory factor with a similar structure to PTH and diverse biological functions. Although PTHrP acts through cell surface receptors, understanding its activity as a growth factor is still controversial (13).
In the current study, the coding sequence of PTHrP “aa 37-177” was isolated and cloned into the pET32a plasmid and then expressed in E. coli host. DNA sequencing and mass spectrometry analysis confirmed the identity of this protein as PTHrP. To the best of authors` knowledge, it was the first study investigating the biological activity of recombinant PTHrP using human breast cancer cell line MCF-7, yet, its role in cell surface receptor-dependent proliferation of breast cancer cell was previously reported by others (9, 10).
Overexpression of PTHrP and PTHrP receptors in breast cancer cells and its role in breast cancer bone metastases (14) suggests that, similar to other growth factors, PTHrP acts through cell-specific receptors. PTHrP induced cell-surface receptors to trigger 2 different G-protein coupled receptors (adenylate cyclase and phospholipase C), which activate protein kinase A (PKA) and protein kinase C (PKC), respectively; this activation initiates different signaling cascades. This signaling system is complicated considering the crosstalk between receptor tyrosine kinase (RTK) and G-protein coupled receptors (reviewed in (15)).
Activation of adenylate cyclase (AC) or phospholipase C (PLC) depends on the concentration of PTHrP and relatively high concentration (micro molar) of PTHrP is required for efficient induction of PLC pathway, in contrast to activation of AC pathway, triggered by physiological concentration (subnanomolar) of PTHrP (16). However, it is also reported that both AC and PLC pathways were activated in PTHrP induced MCF-7 cells (9, 13).
In the current study, 100 nM concentration of PTHrP had the best effect on the cell proliferation of MCF-7 cells (Figure 2). The current study results were consistent with the findings reported by Birch et al., who demonstrated a dose-dependent increase in MCF-7 cell proliferation induced by PTHrP (10). Moreover, Hoey et al., reported an increase in cell proliferation of MCF-7 cells along with an increase in the expression of PTHrP receptor (14). However, others described a decline in cell proliferation rate of PTHrP-treated MCF-7 (9, 13). According to the different and contradictory reports on this matter, the current study designed an additional experiment relying on the anti-hypertrophic effect of PTHrP on in vitro MSC chondrogenesis to verify the sensitivity and reliability of the MCF-7-based method.
There are numerous reports concerning anti-hypertrophic and positive effects of PTHrP on chondrogenic differentiation of MSCs (5, 17). The previous studies demonstrated that application of TGFβ (transforming growth factor β) and BMP (bone morphogenetic proteins) for in vitro chondrogenesis of MSCs leads to induction of hypertrophy in differentiated chondrocytes (18). The study similarly observed an increase in hypertrophy markers (ALP and Col X) using TGFβ1and BMP-2 for chondrogenic differentiation of MSCs. Application of sub-nanomolar concentration (0.1 nM) of recombinant PTHrP for 2 weeks showed a significant decrease in the expression of hypertrophic markers (Figures 4 and 5). In contrast to other reports (9, 13), indicating activation of both AC and PLC pathways in MCF-7 cells induced by submicromolar concentrations of PTHrP, it seems that subnanomolar concentrations of PTHrP have anti-hypertrophic effects on in vitro chondrogenic differentiation through the AC pathway. However, further studies are required to better understand the signal transduction pathway involved in anti-hypertrophy effects of PTHrP.
In conclusion, the current study results indicated that recombinant PTHrP had positive effects on MCF-7 cell proliferation in a certain concentration that can be used as a relatively quick and easy method to analyze its biological activity.