1. Background
According to bioinformatics and microarray analyses, it has been suggested that around 30% of all human genes are regulated by miRNA (1). miRNAs are endogenous 19 - 25 non-coding RNAs involving in the post-transcriptional regulation (2). MicroRNA genes are transcribed by RNA polymerase II, which leads to the formation of the primary transcripts of miRNAs (pri-miRNAs). pri-miRNAs are processed by nuclear RNase III Drosha and develop into a stem-loop structure called miRNA precursor (pre-miRNA). Pre-miRNAs are exported from the nucleus to the cytoplasm via transporter Exportin-5, which is dependent on the GTP-binding nuclear protein Ran (RanGTP). In the cytoplasm, pre-miRNAs are cleaved by a cytoplasmic RNase III named Dicer to form a double-stranded miRNA structure (dsRNA) that is 20 - 25 nucleotides in length. dsRNA is then unwound into two single-stranded mature miRNAs that are incorporated into RNA-induced silencing complex (RISC) (3). The miRNA present in the RISC recognizes a specific sequence motif within 3’-UTR of the target mRNA through seed sequence that contains 7 - 8 nucleotides in its 5’ end and ultimately causes a decrease in the expression of the target mRNA-derived protein (4).
It has been shown in various studies that miRNAs dysregulation is involved in tumor progression (5). Although different approaches are used in CRC treatment including surgery, radiotherapy, and chemotherapy, the risk of tumor recurrence is still the main challenge due to cancer stem cells (CSCs) (6).
CSCs have self-renewal and multi/pluripotency properties and play a crucial role in tumor initiation, metastasis, and resistance to chemotherapy and radiotherapy (7). Colon CSC was discovered in 2007 (8). These cells are characterized by expressing different markers including BMI1 (9). BMI1 is an oncogene that plays a role in cell apoptosis and self-renewal via inhibiting INK4a/ARF that codes two tumor inhibitors named p14 and p16 (10, 11). BMI1 has been identified in several human cancers and is involved in proliferation, invasion, and metastasis as well as in resistance to chemo-radiotherapy (12). Colorectal CSCs survival can be decreased through BMI1 silencing. Few studies have investigated BMI1 inhibition by miRNA in CRC (13).
The aim of this study is to conduct a bioinformatics analysis of various CRC tumor suppressor miRNAs which have potential to target BMI1 and have not been studied at experimental level yet. Tumor-suppressor miRNAs that experimentally have been shown targeting BMI1 in CRC and other types of cancers or those validated in some databases including miRTarbase and TarBase as a suppressor of BMI1 are not discussed in this study.
2. Methods
2.1. Obtaining BMI-1 mRNA Sequence in FASTA Format and miRNA Sequences
The study was approved by the Ethics Committee of Hamedan University of Medical Sciences. BMI1 mRNA sequence with the accession number of NM_005180 and BMI1 3’UTR were obtained from NCBI and UTRdb (www.utrdb.ba.itb.cnr.it), respectively. The mature sequence of all miRNAs was obtained from miRBase (http://www.mirbase.org).
2.2. Bioinformatics Analysis of Potentially BMI-1-Inhibiting miRNAs in CRC
By using several miRNA target prediction tools including TargetScan, DIANA-microT, miRanada, PicTar, miRTAR, mirMAP, miRDB, and FindTar3, the prediction of BMI-1, as a potential target of the known tumor suppressor miRNAs in colorectal cancer, was investigated. The results were scored based on different algorithms for prediction of miRNA pairing to the target sites (especially 3’UTR, because target sites for animal miRNAs are found preferentially in 3’UTR) and free energy (or Gibbs free energy) of binding or secondary structures of 3’UTRs. Finally, the highest-scoring suppressor miRNA was selected. This study began in 2016.
3. Results
3.1. Exploring CRC Tumor Suppressor miRNAs Targeting BMI1 in miRNA Target Prediction Databases
To investigate miRNAs targeting BMI1, eight databases such as TargetScan, DIANA-microT, PicTar, miRanada, mirMAP, miRTAR, miRDB, and FindTar3 were explored. Supplementary File Appendix 1 summarizes the results.
3.2. Analysis of the Data According to TargetScan
Target Scan is capable of predicting the biological targets of miRNAs through exploring the presence of 6, 7, and 8 nucleotides length sequences that are paired to the seed sequence (14). In this database, the targets could be sorted based on PCT (the probability of conserved targeting). PCT ranges from 0 to 1. Higher Pct is related to more specific and more accurate binding of miRNA to its target. In addition, the total context score represents a set of different factors including 3’ UTR length, ORF length complementary base pairing, seed sequence stability, and PCT (15). According to the TargetScan database, miR-330-3p and miR-203 were identified as miRNAs targeting BMI1. The results demonstrated the highest PCT was derived for miR-330-3p that was found to have the highest number of target sites (Table 1).
| miRNA | Conserved Sites | Poorly Conserved Sites | Total Context Score | PCT |
|---|---|---|---|---|
| miR-330-3p | 2 | 2 | -0.34 | N/A |
| miR-203 | 1 | 0 | -0.19 | 0.24 |
Analysis of Results in TargetScan
3.3. Analysis of Data According to DIANA-microT
DIANA-MicroT is used to predict miRNAs targets based on various parameters of miRNA. DIANA-MicroT yields a final score named miTG via integrating data on conserved and non-conserved 3’UTR of the target gene (16). The highest miTG was derived for miR-330-3p (6.4), followed by miR-142-5p (3.1). The highest accuracy that indicates the probability of being a valid target gene for miRNA was estimated for miR-330-3p. Correspondingly, the signal/noise ratio (SNR), indicating the reality of the target gene, was 2.15 for miR-330-3p that is more acceptable than that of miR-142-5p (Table 2).
| miRNA | miTG | Precision of Prediction | SNR |
|---|---|---|---|
| miR-142-5p | 3.16 | 0.08 | 1.09 |
| miR-330-3p | 6.49 | 0.53 | 2.15 |
Analysis of Results in Diana MicroT (SNR: Signal to Noise Ratio)
3.4. Analysis of Results in the PicTar Database
PicTar is an algorithm that is used to identify miRNAs targets. It is determined based on the 3’UTR pairing of the target gene and the seed sequence. The PicTar score was achieved only for miR-330-3p in this database and the scoring level was 6.0151.
3.5. Analysis of Results in the miRDB Database
miRDB is an online database to predict the miRNAs targets. All target scores are derived from thousands of miRNA-target interactions obtained from reliable large-scale experiments. The range of the score is between 50 and 100. miRDB scores over 80 indicate that the predicted target is likely real (17). The results of this database suggested the score of 94 and 82 for miR-152-5p and miR-330-3p, respectively.
3.6. Analysis of Results in miRmap Database
miRmap scores are from 0 to 100 determined based on several thermodynamic and sequence-based parameters including duplex ΔG, evolution, and probability (18). According to miRmap, the highest rate and possible accuracy were attributed to miR-330 (Table 3).
| miRNA | Probability Exact | miRmap Score |
|---|---|---|
| miR-330 | 96.51 | 98.54 |
| miR-520d-5p | 75.05 | 95.10 |
| miR-138-5p | 94.94 | 88.44 |
| miR-328 | 72.29 | 75.29 |
| miR-139 | 34.17 | 73.96 |
Analysis of Results in miRmap (the Results are Arranged by miRmap Score)
3.7. Analysis of Results in miRanada
The mirSVR score in this database is based on the sequence and structural features estimating miRNA effect on the target gene (19). According to the mirSVR algorithm, scores under -0.1 are acceptable; otherwise, the scores are not acceptable. The highest score was derived for miR-203 (Table 4). The lowest rating was given to miR-330-3p among other miRNAs. The PhastCons score of miR-330-3p was higher than the score of the other two miRNAs (Table 4). PhastCons shows the conservation score ranging between 0 and 1.
| miRNA | mirSVR | PhastCons Score |
|---|---|---|
| miR-203 | -0.9411 | 0.7238 |
| miR-212 | -0.9321 | 0.6761 |
| miR-330-3p | -0.1101 | 0.7312 |
Analysis of Results in miRanda (the Results are Arranged by miRSVR Score)
3.8. Analysis of Results in the miRTar Database
miRTar predicts the interaction between miRNA and the target based on various factors including minimal free energy (20). In this database, the default threshold for least amount of energy and the total score are considered ≤ -14 and ≥ 140, respectively. Accordingly, BMI1 was found to be the predicted target of miR-520d-5p, miR-145, miR-198, miR-330-3p, and miR-203. To compare and rank miRNAs more strictly, the threshold levels for minimal free energy and total score were changed to ≤ -18 and ≥ 160, respectively, and then miR-330-3p was found to be the only miRNA targeting BMI1.
3.9. Analysis of Results in the FindTar3 Database
FindTar3 ranks the targets based on certain criteria such as miRNA-target base pairing and duplex energy of miRNA-target. In this database, the main point is the relative pairing of miRNA to its target, according to which the loop score is determined. If this score lies between 15 and 20, the predicted target score will be acceptable while those with a score higher than 20 are considered optimal target (21). Among the various scores obtained in this database, miRNAs with the highest scores were selected and listed in the Supplementary File Appendix 1.
4. Discussion
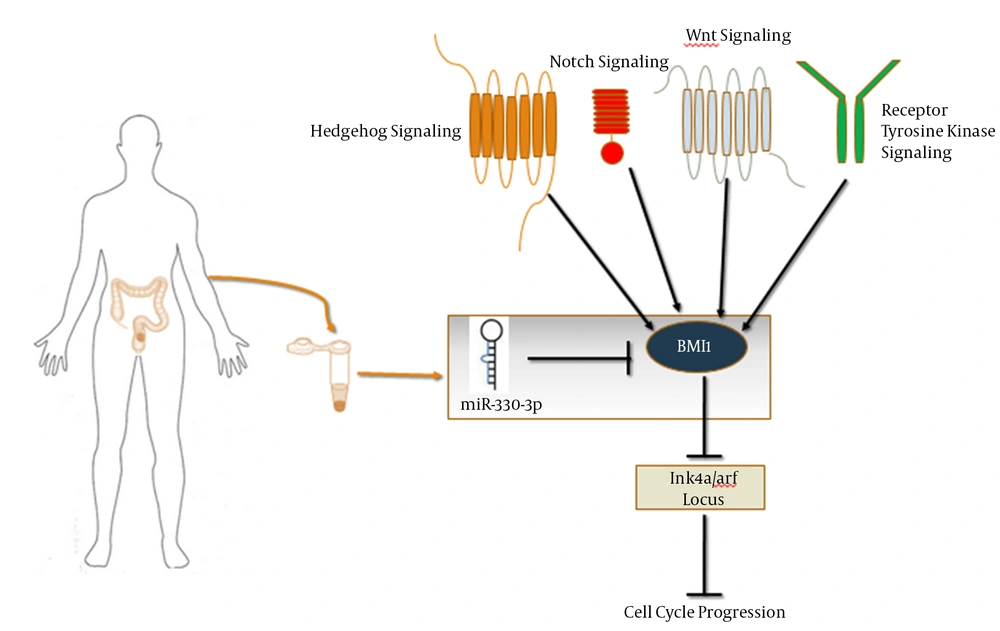
The CSCs are a subpopulation of tumor cells that share certain similar characteristics with normal stem cells and are responsible for initiating and maintaining tumorigenesis and its recurrence (22). CSCs can serve as appropriate therapeutic targets in cancer targeted therapy. Many pieces of evidence have indicated that BMI1 plays a significant role in proliferation, self-renewal, and differentiation of several types of stem cells and precursors cells (23). BMI1 is required for not only self-renewal of different types of tissue-specific stem cells but also the proliferation of cancer cells (24). As an epigenetic inhibitor, BMI1 inhibits Ink4a/ARF locus that codes P16Ink4a and P19ARF (the human homologue of P14ARF) via interaction with other components of PRC1 (25) (Figure 1). P16Ink4a and P19ARF are the upstream activators of Rb and P53 pathways, respectively (26).
Many studies have demonstrated the increased expression of BMI1 at the mRNA and protein levels in CRC (27). BMI1 mediates ubiquitination of histone and is essential for proliferation of colon cancer in vitro and in vivo (28). It has been suggested that BMI1 plays a central role in maintaining colorectal cancer cells growth and tumor progression through regulation of self-renewal (29). MiRNAs are considered key factors in cancer genetics and potentially can be used as anti-cancer drugs through their transfer to the cell (30). In addition, miRNAs should be considered as promising targets for cancer diagnosis and treatments monitoring (31). Although different techniques such as microarray are available to detect the miRNAs’ target genes, bioinformatics analysis is an inexpensive and convenient approach to conduct such studies. To the best of our knowledge, it has been shown that only miR-218 and miR-508-3p may inhibit BMI1 at the RNA or protein level in CRC (32, 33).
Therefore, considering the significance of BMI1 in CSCs and the potential association between this molecule and different miRNAs in cancer, different bioinformatics tools were used to find miRNAs that suppress BMI1. To achieve this purpose, the known tumor-suppressing miRNAs in CRC were selected as candidates in bioinformatics analysis so that experimental investigations can be conducted to gain a better understanding of colorectal carcinogenesis. Besides, the importance of miRNAs as a potential biomarker in the study of colorectal cancer has been well demonstrated. According to the results of bioinformatics analysis in the current study, out of the investigated miRNAs, the highest score for targeting BMI1, as one of the markers of CSCs, was obtained for miR-330-3p. According to all databases used in this study, only miR-330-3p was found to target BMI1.
Most databases that were investigated in this study (apart from miRDB, miRanda, and FindTar3) gave the highest score for miR-330-3p. However, the score of this miRNA was acceptable in miRDB, miRanda and FindTar3 databases, as well. These high scores represent the stability of the seed sequence binding of miR-330-3p to 3’UTR of the BMI1 gene and high probability of targeting. Due to different algorithms and scoring schemes, some differences in the scores were noted among some databases; altogether, miR-330-3p was found to have the highest score. The expression of miR-330-3p, as a tumor suppressor, has been reported to decrease considerably in CRC tissues and cell lines (34). Yan Li et al. study demonstrated that miR-330-3p induces apoptosis and inhibits proliferation via inhibiting cdc42 (35).
The inhibitory role of miR-330-3p in gastric cancer was demonstrated by inhibiting Musashi-1 (MSI1) (36). MSI1 is the marker of normal and cancer stem cells (37). Therefore, given that BMI1 is the marker of colon cancer stem cells, the decreased expression of miR-330-3p is likely to be associated with the activation of colon cancer stem cells signaling pathways. According to the findings of the current study, the association between miR-330-3p and BMI1 expression can be investigated in experimental studies in order to evaluate its applicability as a biomarker in CRC (Figure 1). Since the serum level of BMI1 increases in CRC patients (38) and the miRNAs in the serum or plasma have been recently reported to be appropriate biomarkers with high specificity and sensitivity (39), the simultaneous study of these two molecules in the serum or plasma of colorectal cancer patients can help achieve a double non-invasive biomarker panel.
4.1. Conclusion
Bioinformatics tools are one of the fastest and inexpensive approaches to find potential miRNAs targets. The results of the bioinformatics analysis of this study have shown the highest prediction scores for miR-330-3p in most miRNA-target prediction databases. BMI1 is involved in the function and survival of CSC; hence, the suppression of BMI1 expression with miR-330-3p would be not only a favorite therapeutic target in CRC targeted therapy but also valuable in a better understanding of the molecular mechanisms in CSC self-renewal and survival. Moreover, the expression evaluation of miR-330-3p and BMI1 in serum/plasma or tissues of CRC patients would be useful in biomarker studies. Altogether, the experimental validation of an association between BMI1 and miR-330-3p via techniques such as qRT-PCR, western blot, and luciferase assay is suggested.