1. Background
Urinary pathogen Escherichia coli (UPEC) are the first cause of community-acquired and responsible for 50% of hospital-acquired urinary tract infections (UTIs) (1, 2). UPEC strains act as opportunistic intracellular pathogens that colonize the urinary tract following predisposing factor including kidney stone, impaired immune function, kidney injury, history of urinary tract surgery, pregnancy, tumor, congenital malformation, diabetes, urinary retention, tuberculosis, or urinary tract manipulations such as catheterization (3). UTIs are the predominant type of bacterial infection diagnosed nowadays. In fact, E. coli are responsible for more than 85% of all UTIs controlled using antibiotic therapy all over the word.
Antibiotics are considered as the first-line treatment for bacterial infections that excessive and inappropriate use of them result in antibiotic resistance, which is now a rapidly growing global problem (4, 5). E. coli is one of the opportunistic pathogens in hospitals, especially in inpatients (6). In addition to UTIs, these bacteria cause many nosocomial infections such as sepsis, ulcers, gastroenteritis, and neonatal meningitis. E. coli is resistant to β-lactam antibiotics due to the acquisition of plasmids encoding broad-spectrum β-lactamases (7). The updated classification for β-lactamases includes group 1 cephalosporinases; group 2 broad-spectrum, inhibitor-resistant, and extended-spectrum β-lactamases, and serine carbapenemases; and group 3 metallo-β-lactamases (8). Extended-spectrum β-lactamases (ESBLs) have particular importance in antimicrobial therapy and have the ability to hydrolyze the oxyimino-β-lactams such as cefotaxime, ceftazidime, ceftriaxone, or cefepime (9). To date, more than 350 different natural ESBL variants are known classified into nine distinct families that the main ones are the TEM and SHV groups and CTX-M derivatives; in addition, IMP gene is also found in ESBL classes (10). ESBLs are often reported in E. coli and Klebsiella spp., but are also detected in other Enterobacteriaceae (11). In addition, ESBLs exhibit co-resistance to many other classes of antibiotics, resulting in the limitation of the therapeutic options (9). Hence, bacteria producing broad-spectrum β-lactamases such as E. coli are clinically important since they confer a broad spectrum of drug resistance patterns and increase mortality rate, especially in patients admitted to intensive care units (12, 13). The prevalence of ESBL-producing E. coli increased from 33.7% in 2005 to 60% in 2009 (14). In the study by Boisen, out of 224 E. coli isolated strains, 116 samples (47.1%) were resistant to ceftazidime and 96 (39.2%) to cefoxitin, of which 109 strains were ESBL-positive. Also, TEM, CTX-M, and SHV genes were positive in 87.1%, 68.8%, and 70.6% of the isolates, respectively (15). Soltan Dallal et al. in a study from Tehran, Iran showed that of the 200 isolated E. coli, 128 strains were ESBL- producing, 99 of which harbored the CTX-M gene, and seven (5.5%) carriers the SHV gene (16). In the study by Shahcheraghi et al. 93.6% of the strains produced ESBL enzymes inducing resistant against piperacillin, while resistance to piperacillin-tazobactam was observed in 26.6% of the patients (17). In the study by Haghighat Panah et al. the results of susceptibility to antibiotics in different strains showed that the highest rate of resistance was to amoxicillin and all isolates were susceptible to imipenem, and the TEM gene detected in 32.5% of the strains had a direct relationship with resistance to antibiotics (18).
Since resistance to antibiotics is increasing worldwide, especially in the developing countries, therefore, further studies are needed to determine the prevalence of resistance to antibiotics in E. coli and the necessity of using appropriate tools for infection control and optimal therapeutic strategies to reduce the prevalence of such bacteria. Some antibiotics such as ceftriaxone, imipenem, and piperacillin did not promote the colonization of ESBL-positive strains thus far and therefore, they are commonly administered to treat UTI. Hence, these antibiotics were evaluated in the present study.
2. Objectives
The current study aimed at determining the frequency of SHV, CTX-M, and IMP genes and their role in the emergence of resistance to ceftriaxone, imipenem, and piperacillin in E. coli isolated from UTI.
3. Methods
In the current descriptive-analytical and cross sectional study, according to cases with UTI referent to hospital, 3000 urine samples of outpatients and inpatients from March 2014 to the end of July 2015 were investigated. The sample size was determined based on the average number of urine samples referring to the Laboratory of Velayat Hospital (Damghan, Semnan province, Iran) for five months. E. coli strains were isolated from 270 samples after culturing the urine and applying standard microbiology methods. Before the experiment, each subject signed the informed consent form.
3.1. Culture
All specimens were inoculated into blood agar and MacConkey agar, and then incubated at 37°C. Bacterial isolates were verified by colony morphology, staining, motility, and biochemical tests.
3.2. Disc Diffusion
The screening test was performed using disc diffusion method according to the CLSI (The Clinical and Laboratory Standards Institute) guidelines. Gram-negative bacteria isolated from culture media were considered as positive, which resulted in the formation of an inhibition zone around antimicrobial discs including 30 µg of ceftriaxone, imipenem, piperacillin, and piperacillin-tazobactam.
3.3. Double-Disc Synergy Test
ESBL production of the isolated Gram-negative bacteria was investigated by the double-disc test. The bacteria considered ESBL-positive if the inhibition zone around ceftriaxone, imipenem, and piperacillin discs on Muller-Hinton agar plus 10 µg clavulanic acid were ≥ 5 mm in diameter.
3.4. Extracting DNA from Bacteria
DNA extraction was performed using boiling method, according to the protocol described by Antunes et al. (19) with some modifications. Firstly, a colony taken from the Tryptone Soy Agar was inoculated into Luria broth medium and then incubated at 37°C for 18 - 24 hours; 1000 µL of culture medium containing the bacteria was centrifuged at 1300 - 1500 rpm for 10 minutes at room temperature (RT). The supernatant was discarded and 500 µL of the TE buffer (10 Tris mM and 1 mM ethylenediaminetetraacetic acid) was added to the sediment and vortexed to form a homogenous suspension. The Eppendorf tubes were again centrifuged at 1300 rpm for 10 minutes; the resultant supernatant was discarded and then re-centrifuged at 1300 rpm. Final sediment was dissolved in 500 µL of TE RNase buffer and heated up to 100°C for 13 minutes. The samples were centrifuged at full speed (13,000 - 15,000 rpm) for 20 minutes at RT. The supernatant was aspirated by gently pipetting and DNA concentration was estimated using a UV spectrophotometer (50 ng/µL). The extracted DNA was stored at -10°C. DNA absorbance was measured and calculated at a 260/280 nm ratio to evaluate the DNA quality. Likewise, 2 µL of extracted DNA was placed in NanoDrop (Thermo, 2000) and the UV absorptions at 230, 260, and 280 nm were measured and 260/230 and 260/280 nm ratios were calculated. In order to qualitatively evaluate DNA, 1% agarose gel and fluorescence staining for DNA was used. Samples with distinct bands and no defects and smears were used for subsequent analyzes.
3.5. Polymerase Chain Reaction
To perform the polymerase chain reaction (PCR), primer sequences were extracted from the studies and examined using the Blast program, and then the most suitable primer sequence was selected. Primers related to CTX-M, SHV, and IMP genes were used according to the following Table 1.
| Amplicon Size, bp | Sequence of Primer (5’ → 3’) | Primer Name |
|---|---|---|
| 800 | CTX-M | |
| F | GTTACAATGTGTGAGAAGCA | |
| R | CCGTTTCCGCTATTACAAAC | |
| 885 | SHV | |
| F | CACTCAAGGATGTATTGTG | |
| R | TTAGCGTTGCCAGTGCTCG | |
| 187 | IMP | |
| F | GGA ATA GAG TGG CTT AAT TCT | |
| R | CCA AAC TAC TAG GTT ATC T |
The primers were diluted according to the manufacturer’s instructions (Cinnagen company, Iran), and then the final concentration was determined at the nanogram levels by a spectrophotometer. To determine the optimum temperature for annealing the primers, the gradient PCR was used with temperatures ranging from 57°C to 65°C (57, 57.7, 58.4, 59.1, 59.9, 60.6, 61.3, 62, 62.8, 63.5, 64.2, 65°C). For the SHV gene, the annealing temperature was 55°C. Also, the process was performed in a thermocycler machine in 30 cycles, and PCR products were repeated using agarose gel electrophoresis and a higher volume of DNA.
3.6. Qualitative PCR Product Analysis
A 10-µL of each PCR product was mixed with 2 µL loading buffer and poured into the 1% gel well and a 1.5-µL DNA ladder was poured into the first well. Also, the second and other wells were PCR products with the positive and negative controls. After loading, negative and positive electrodes were connected to the electrophoresis tank. Then, the electric current was applied at 80 - 100 V for 60 minutes. When the samples were migrated more than two-thirds of the length of the gel, the gel was placed in transilluminator under UV light and the position of the bands was investigated by ultraviolet radiation.
3.7. Phenotypic Confirmatory Test
For this purpose, disc diffusion test was performed on the Muller-Hinton agar.
3.8. Data Analysis
All the data analyses were performed in triplicate by ANOVA with SPSS version 18 (Chicago, IL, USA), and using descriptive statistics (mean, standard deviation, and absolute and relative frequency and frequency distribution).
4. Results
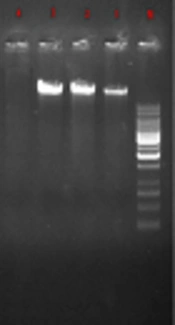
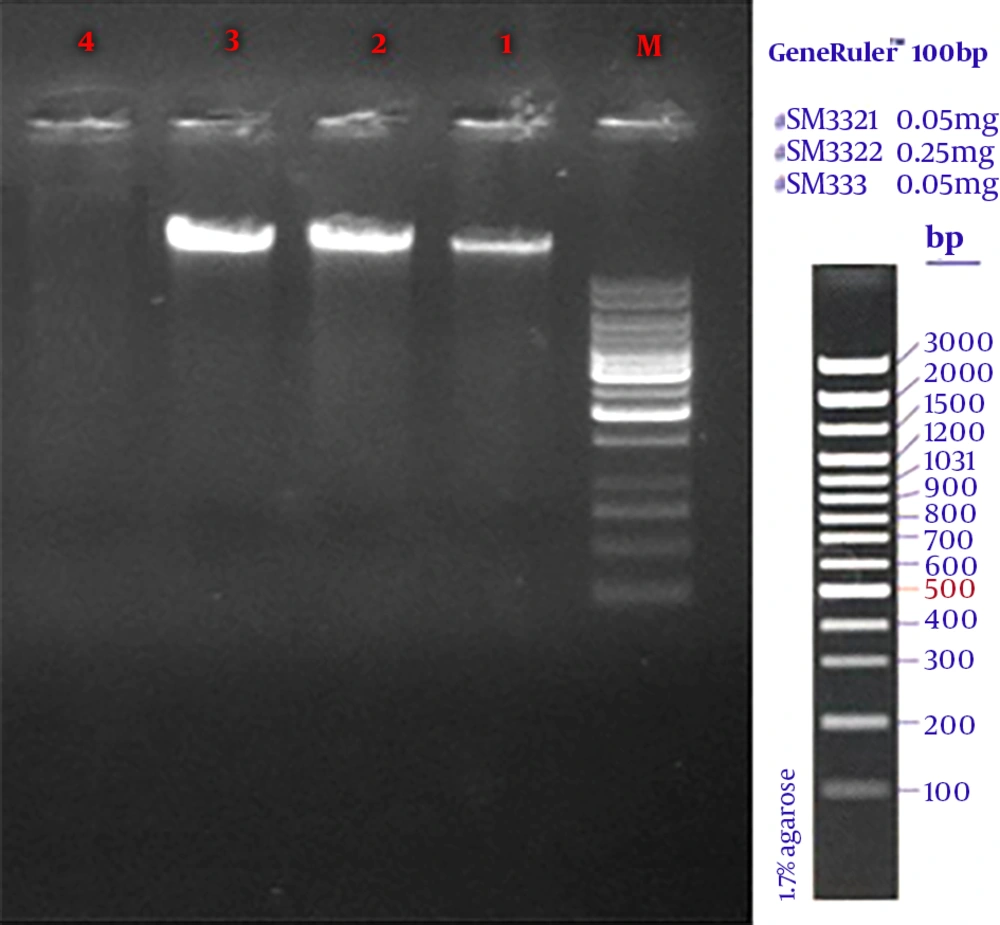
In the current study, 270 samples were identified as E. coli using phenotypic tests. Of the 270 isolated E. coli, 120 strains (44.4%) were ESBL-positive and 150 (55.56%) ESBL-negative. Likewise, of the 270 E. coli strains, 36 (28.33%) were isolated from males and 84 (71.67%) from females. Also, most subjects (24%) were within the age range of 20 to 30 years. PCR results showed that among the ESBL-positive samples, 36 strains (30%) harbored CTX-M and 33 (27.5%) SHV genes (Table 2). The amplified product was resolved by electrophoresis on gel agarose; in Figure 1, a 1080-bp band confirmed the presence of a β-lactamase gene in the bacteria. No IMP gene was detected in the tested samples.
| Bacteria | ESBL-Producing Bacteria | CTX-M Positive | SHV Positive | IMP Positive | |
|---|---|---|---|---|---|
| Positive | Negative | ||||
| E. coli | 44.44 | 55.56 | 30 | 27.5 | 0 |
aValues are expressed as percentage.
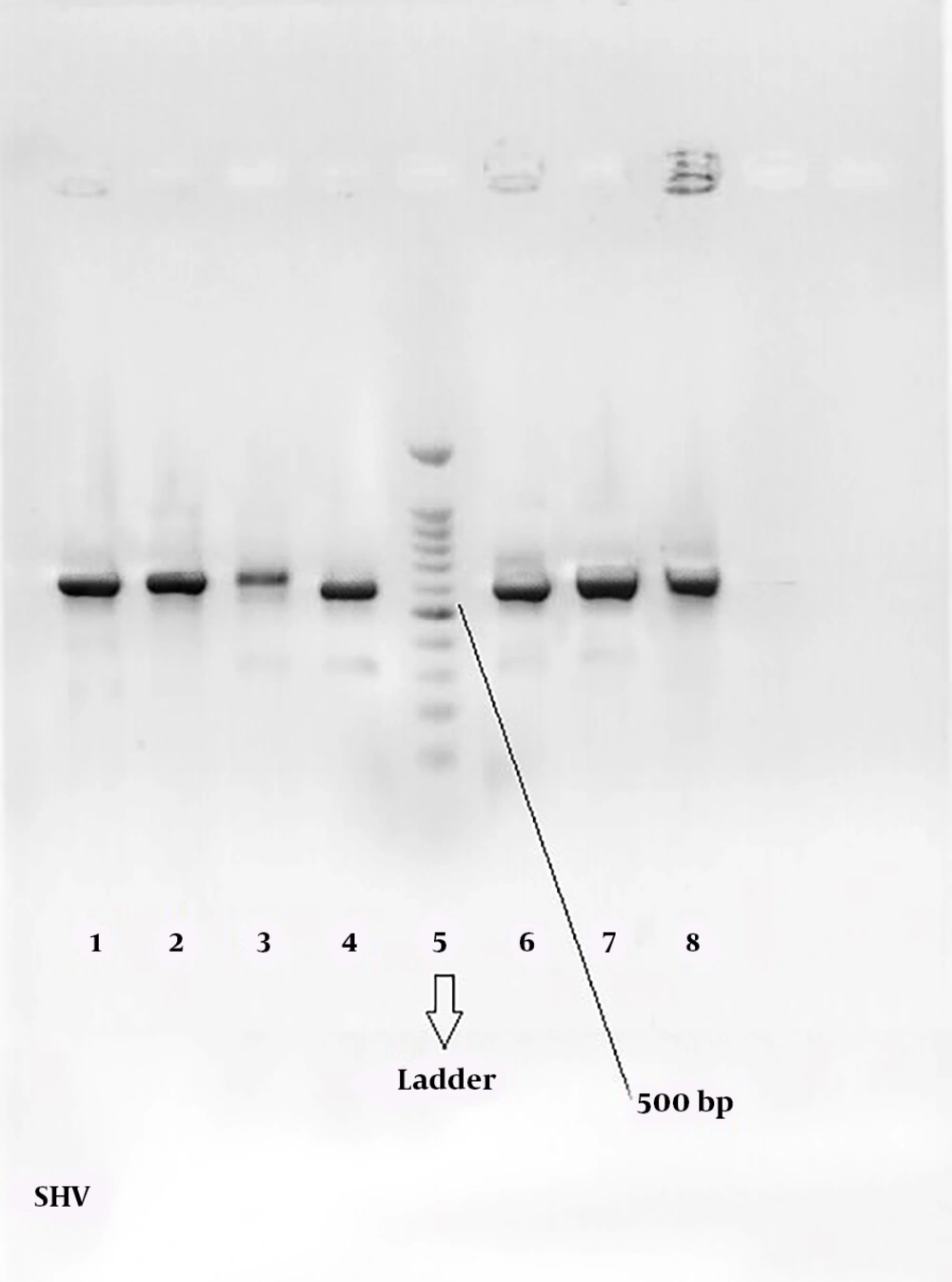
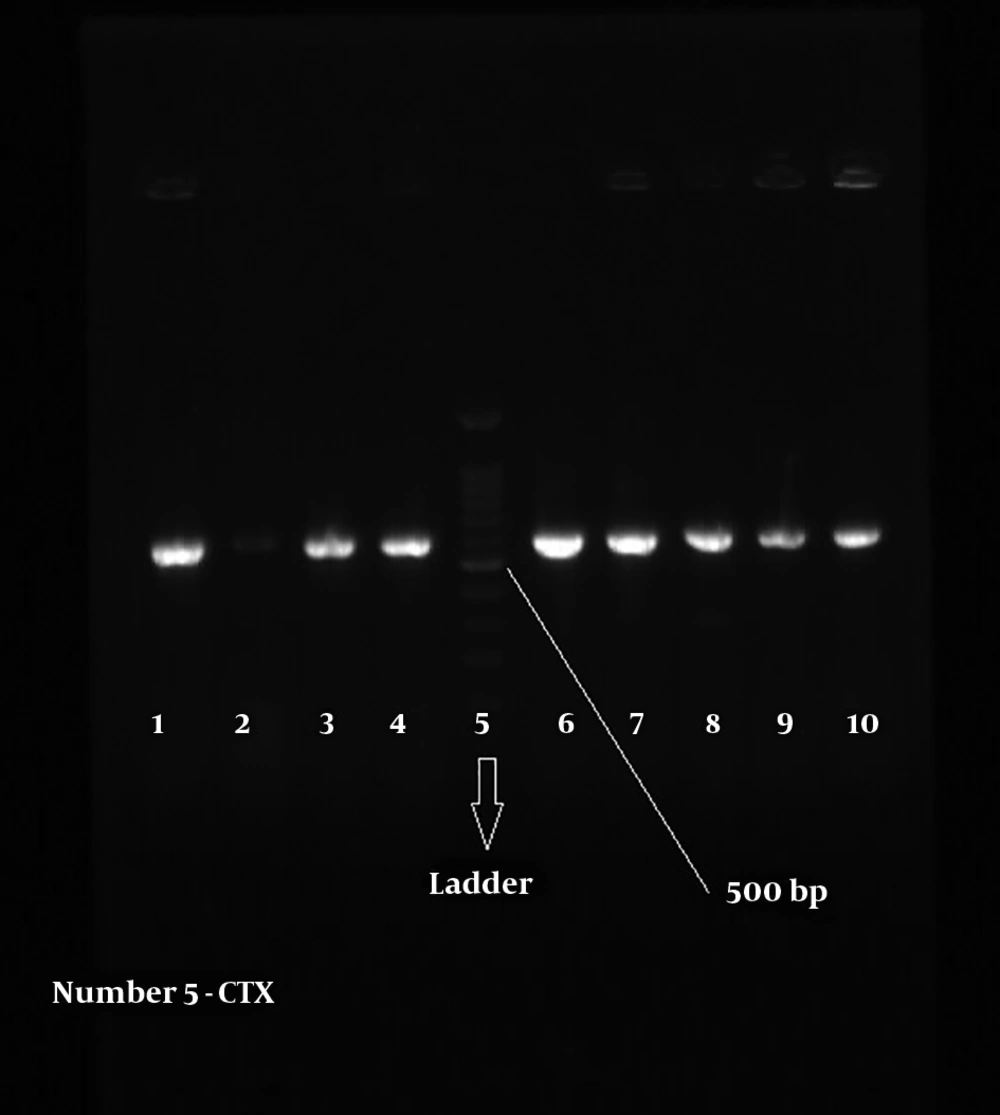
Figures 2 and 3 show the 568-bp band produced by the amplification of a segment of SHV and CTX-M genes in ESBL-positive samples, respectively. Totally, 71.67% of the patients with UTI were female and 28.33% male; therefore, there was a significant difference (P = 0.039) in terms of gender among patients with UTI in the hospital.
Based on antibiogram data, the resistance to ceftriaxone, piperacillin and imipenem was different among the isolated bacteria. As shown in Table 3, the highest resistance in the isolated strains (100%) was against piperacillin both in male and female patients. However, the frequency of resistance to ceftriaxone was more than imipenem in all samples. Regarding gender differences in terms of response to treatment, resistance to ceftriaxone were mostly observed in male patients (65.63%) compared to females (58.33%).
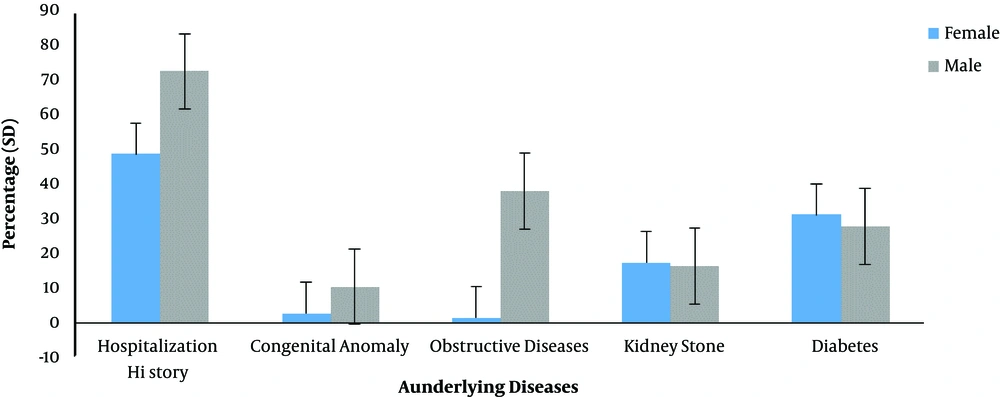
As it can be observed in Figure 4, the distribution of underlying diseases including diabetes, kidney stones, obstructive disease, congenital anomalies, and hospitalization history was different in patients with UTI. The hospitalization history had the highest rate of incidence in all the studied cases. Also, the prevalence of diabetes was 31.08% in female and 27.59% in male patients with UTI.
| Gender/Response | Ceftriaxone | Imipenem | Piperacillin |
|---|---|---|---|
| Female | |||
| Resistant | 49 (58.33)Ab | 31 (36.47)B | 85 (100)A |
| Intermediate susceptible | 1 (1.19)C | 8 (9.41)C | 0 B |
| Susceptible | 34 (40.48)B | 46 (54.12)A | 0 B |
| Male | |||
| Resistant | 21 (65.63)A | 42 (36.67)B | 33 (100)A |
| Intermediate susceptible | 0 (0)C | 11 (10)C | 0 B |
| Susceptible | 11 (34.38)B | 62 (53.33)A | 0 B |
aValues are expressed as No. (%).
bThe same letter in each column for male and female patients indicates no significant difference at P = 0.039.
5. Discussion
In the current study, the frequency of ESBL-producing strains and the role of CTX-M, SHV, and IMP enzymes in the emergence of resistance against ceftriaxone, imipenem and piperacillin in E. coli strains isolated from UTI samples were investigated. A significant number of samples were ESBL-positive and harbored CTX-M and SHV genes. Regarding the underlying diseases, considerable differences were observed in the studied patients.
According to the results, 44.44% of E. coli strains were ESBL-producing bacteria. While, in the study by Ling et al. in China, the rate of ESBL production in E. coli strains was 16% (20). In the study by Duttaroy et al. in India performed on 187 strains of E. coli and Klebsiella sp., 53 isolates (29.1%) were ESBL-producing (21). Also, among the ESBL-producing Enterobacteriaceae strains isolated from a hospital in France, a significant percentage of the isolated bacteria were E. coli (40.5%) (22). Therefore, it seems that the prevalence of ESBL-producing E. coli is relatively high in hospitals of Iran, which may be the cause of the high rate of resistance to antibiotics. Among the ESBL-positive samples, 36 strains (30%) harbored the CTX-M gene. The prevalence of CTX-M gene in the current study isolated strains was similar to that of the studies from other countries. In a study from South Korea (2004) on 603 Enterobacteriaceae strains, 6.8% of the total samples and 44.1% of the resistant samples carried the CTX-M gene (23). In the study by Eisner et al. from Austria, 58% of the examined E. coli strains harbored CTX-M gene (24). In the studies by Mobin et al. (25) and Samadi et al. (26) CTX-M gene was detected in 25.1% and 75% of the ESBL-producing E. coli, respectively. Mirzaee et al. examined 160 isolates of E. coli to investigate the expression of CTX-M β-lactamases by PCR and reported that 37.8% of the samples were CTX-M-positive (27). A reason for the difference in their results with those of the current study may be different origins of the collected samples and different strategies to control the resistance to antibiotics. Furthermore, 33 samples (27.5%) harbored SHV gene. In a study conducted on 343 strains of Enterobacteriaceae in Budapest, 252 isolates produced ESBL, of which 11.5% were E. coli that 92.1% carried the blah SHV gene. The study showed that the prevalence of the SHV gene was higher than the other β-lactamase genes (28). In the current study samples, the SHV enzyme had the highest prevalence. In the study by Sharma et al. 100 isolates of E. coli were investigated in the Indian hospitals, 56% of them carried TEM gene and 60% the SHV gene (29), while Shahcharaghi et al. identified the SHV gene in 6% of ESBL-producing E. coli (17). According to the obtained results, the IMP gene played no role in antibiotic resistance of E. coli strains isolated from UTI samples collected in a hospital in Damghan. As a matter of fact, 70 (60.34%) ESBL-positive samples showed resistance to ceftriaxone; also, imipenem resistance was observed in 42 (36.52%), and all showed complete resistance to piperacillin.
Extensive β-lactamases are often plasmids, and since these plasmids are easily transmitted among different types of bacteria, particularly in the Enterobacteriaceae family, the combination of resistant genes leads to the formation of strains with multiple-drug resistance. Since plasmids have an independent chromosomal replication and their replication is not dependent on cell and chromosome divisions, they replicate a lot at the cell and, if transmitted to other strains, antibiotic resistance arises (5, 7). In the study by Nowroozi et al. the most effective antibiotic against the isolated E. coli was imipenem (7.93%) (30). In the study by Asadpour Rahimabadi et al. among the isolated E. coli, 36.92% were ESBL-producing strains and imipenem was the most effective antibiotic (31). Also, in the study by Mirsalehian et al. imipenem-resistance was detected in 36.53% of the samples, which was the lowest resistance rate among antibiotics (32). In these studies, similar to the current study, imipenem was the most effective antibiotic against E. coli. However, a significant number of the samples were resistant to antibiotics, which may indicate indiscriminate administration and overprescription of antibiotics, especially in hospitals.
The presence of four underlying conditions including diabetes, kidney stones, obstructive disease, and congenital anomalies as well as hospitalization history in patients with UTI was investigated. According to the results (Figure 1), the percentage of kidney stones and diabetes was higher in female patients with UTI than males. By contrast, obstructive disease and congenital anomalies were higher in male patients than females. Therefore, diseases such as kidney stones and diabetes seem to play an important role in the development of UTI. Hence, the control of these diseases is very effective in preventing UTI and ultimately preventing antibiotic resistance. Also, according to the results, the rate of hospital admission was higher in male patients and since UTI was more common in female patients, it can be concluded that the history of hospitalization plays a small role in the development of UTI-causing E-coli.
The current study findings indicated that the resistance to extended-spectrum cephalosporins including ceftriaxone and imipenem were different in bacteria colonized urinary tract of male and female patients. The bacteria isolated from male patients were more resistant to ceftriaxone and imipenem. Furthermore, all the isolated strains were 100% resistant to piperacillin.
The prevalence of Enterobacteriaceae bacteria is different from country to country and from hospital to hospital. In fact, different results can be one of the limitations of this type of studies. However, the duration of antibiotic therapy, the overall evaluation of case and control groups regarding deface factors were the potential bias of the study.
These findings also showed the importance of E. coli in the hydrolysis of β-lactam drugs, especially extended-spectrum cephalosporins. Therefore, it is recommended to consider the detection of these genes in E. coli strains isolated from clinical specimens in microbiological laboratories.
5.1. Conclusions
The current study results demonstrated the presence of CTX-M-, SHV-, and IMP-type ESBLs in Iran and indicated the important role of resistance to β-lactam antibiotics worldwide. Resistance to β-lactam agents and third-generation cephalosporins is a serious and growing concern. The findings of such studies emphasize providing practical approaches to prescribe drugs, necessity of equipping laboratories with phenotypic diagnostic methods and molecular techniques to identify and determine the type of genetic resistance, estimation of the prevalence of resistant bacteria, and application of the necessary measures to treat patients and control the resistance of the bacteria. Furthermore, the most effective treatments should be administered to prevent the emergence of antimicrobial-resistant bacteria.