1. Background
Kidney stone disease affects approximately 1 - 19.1% of the world population (1, 2). The disease has been described to have accompanying severe pain, and its treatment is a financial burden on the healthcare system. Various risk factors can exacerbate the formation or recurrence of calcium nephrolithiasis (3). As the most common metabolic abnormality, hyperoxaluria increases the risk of calcium stone formation (2, 4). Urine oxalate is obtained from the intestinal absorption of dietary oxalate and the endogenous metabolism of various precursors, mainly ascorbic acid and glyoxylate (4). Dietary oxalate absorption can be affected by several factors, among which intestinal microbes have received much attention (5).
Oxalobacter formigenes is part of the bacterial flora found in the human colon and many other mammalian species and is unique in that it requires oxalate as a source of energy and carbon (2). A role for this organism in stone diseases has been considered since the discovery of O. formigenes in 1985 and the recognition that it lives in the human intestine and degrades oxalate (5). Preliminary case-control studies with a small number of individuals have shown that colonization with this bacterium may provide protection against nephrolithiasis (5). Colonization of O. formigenes in the gut by polymerase chain reaction (PCR) has been proven in current studies (6, 7). However, the exact amount of these colonizations cannot be measured. Accordingly, qPCR enables us to quantify the rate of colonization (8, 9).
The measure of urinary oxalate excretion in colonized cases was lower than in non-colonized individuals, despite the high variability in oxalate excretion and lack of control of oxalate and calcium in the diet collected in the urine. However, 24-hour urine is the best available tool to assess the contribution of dietary oxalate to urinary oxalate excretion (10).
In general, studies on the degradation of oxalate in the gut by microorganisms have focused on O. formigenes, and it has been reported that colonization with this organism reduces the risk of recurrent calcium oxalate (CaOx) nephrolithiasis formation by 70% (11). Formaldehyde coenzyme transferase A (FCR) and oxalyl coenzyme A decarboxylase (OXC) play a key role in oxalate degradation by O. formigenes (12, 13). The use of oxalate in anaerobic culture media is a practical diagnostic strategy to identify this bacterium. However, due to the presence of similar oxalate-degrading bacteria in the intestine and different strains of Oxalobacter, this method is not highly specific and sensitive. In this regard, molecular methods such as polymerase chain reaction (PCR), quantitative real-time PCR (qRT-PCR), and Southern blot, using specific DNA fragments, can be replaced with previous methods by increasing the sensitivity and specificity in identifying this bacterium (14). Owing to the necessity for new treatment methods to prevent kidney stones, treatment with Oxalobacter may be an effective approach in patients with hyperoxaluria and also to prevent the formation of kidney stones (15).
2. Objectives
This study aimed to evaluate the prevalence of O. formigenes in patients with CaOx kidney stones (with and without hyperoxaluria) compared with the normal control group. The relationship between the presence of CaOx and oxalate in the study population was also evaluated.
3. Methods
3.1. Study Design and Ethical Considerations
In this case-control study, urine and fecal samples of 125 patients were evaluated with informed consent, of which 73 samples were from patients with CaOx kidney stones, and 52 samples were from healthy individuals. The existence of kidney oxalate stones was clinically confirmed by kidney and urinary tract physicians in all subjects included in this study. This study was approved by the ethics committee of Tehran Science and Research Branch, Islamic Azad University (IR.IAU.TNB.REC.1399.025).
3.2. Bioinformatics Analysis and Primer Design
The different gene regions of O. formigenes were evaluated through bioinformatics software (CLC main workbench 12, Gene Runner, Alel ID). Next, the two gene regions FRC and OXC, were selected, and specific primers were designed. The specificity of the primers was confirmed by examining their similarity to other bacterial sequences registered in the Gene Bank (http://www.ncbi.nlm.nih.gov/BLAST).
3.3. Biochemical Analysis of Urine and DNA Extraction from Feces
A 24-hour urine sample was collected to examine excretory oxalate of patients with renal CaOx stones. Samples were evaluated using oxalate and citrate kits by Enzymatic/Endpoint-colorimetric method (Byrex Fars, Fars, Iran). Oxalobacter DNA was extracted using a DNA extraction kit from feces (Favorgen Biotech Corporation, Taiwan), according to the manufacturer’s instructions. A positive control sample was extracted simultaneously with the other samples.
3.4. Setting up PCR and qRT-PCR
The FRC and OXC regions, corresponding to extracted DNA, were amplified by two pairs primers, FRC.F: 5'CCAGCCTACAACACATTCG3', FRC.R: 5'TCAGAACTTCGTCGGTATGT3' and OXC.F: 5 'AATCTAGAGTTGACTGA3', OXC.R: 5 'TTGATGCTGTTGATACG3' using thermocycler (Applied Biosystems, CA, USA), including 12 µL PCR reaction mixture (Amplicon, Denmark), 0.8 µL forward primer (FRC.F or OXC.F), 0.8 μL of reverse primer (FRC.R or OXC.R), 8.15 μL of distilled water and 3 μL of DNA. The temperature protocol includes a primary denaturation at 95°C for 5 minutes followed by 40 cycles: 94°C for 45 seconds (Denaturation), 56°C for 20 seconds (Annealing), 72°C for 30 seconds (Extension), and a cycle of 72°C for 5 minutes. The final product was electrophoresed on 1.5% agarose gel and evaluated by gel docking device (Cambridge, Warwickshire, UK).
Amplification of FRC and OXC by qPCR assay was performed on Step One PlusTM (Applied Biosystem, CA, USA) in the final volume of 12.5 μL including 0.5 μL of each primer (10 Pmol / μL), 2.5 μL of HOT FIREPol® EvaGreen® qPCR Mix Plus (Solis Bio Byne, Estonia) and 2 μL of DNA or PCR product. First, a temperature of 95°C was provided for 15 minutes to start and activate the enzyme. Denaturation process was performed at 95°C for 30 seconds, annealing at 56° C for 30 seconds, and extension at 72°C for 40 seconds in 40 cycles.
3.5. Statistical Analysis
In this study, medicals statistical software (version 19.0.5) was used to calculate and compare the sensitivity and specificity of selected gene regions and the techniques. Using Kolmogorov-Smirnov and Shapiro-Wilk statistical tests, 125 samples were also examined for the normality of the statistical population.
4. Results
4.1. General Findings
In this study, urine and fecal samples of 125 people who were referred to medical diagnostic laboratories were evaluated in terms of the amount of oxalate and the presence of Oxalobacter. Among the studied samples, 52 samples did not contain oxalate stones, and 73 samples had oxalate stones. The presence or absence of stones was determined by a physician's examination and based on the results of clinical tests and ultrasound. The age range of the subjects was between 1 and 73 years, and it was reported that people with an average age of 41.98 years old had no stones, and the ones with an average age of 29.27 years old had stones. Women and men composed 68% and 32% of the population, respectively. However, P-value = 0.177 showed no significant relationship between sex and the incidence of CaOx stones. According to Table 1, the patients were divided into three groups in terms of age: under 20 years (y), between 20 to 35 yrs, and over 35 yrs. People over 35 who constitute more than half of the people in our study (57.6%), make up 47.94% and 71.15% of people with CaOx stones (n = 73) and without oxalate stones (n = 52), respectively. Although most people with and without stones were over 30 years, the group under 20 years, which was 26.4% of the population, comprised 35.61% (26/73) of those who had oxalate stones. The results of the relationship between age groups with the presence or absence of oxalate stones showed a significant relationship between these two factors (P-value = 0.004). In order to evaluate the amount of oxalate excretion, the 24-hour urine sample of the study population was examined. The rate of oxalate excretion in people with stones was 22.93 mg/24h, and in people, without stones it was 30.17 mg/24 h.
The results showed that out of 14 people who had oxalate excretion below 10 mg/24 h, 11 (78.6%) subjects had CaOx stones. After that, most patients with oxalate stones were in the group whose oxalate excretion was in the range 10 - 25 mg/24 h (66.1%), while only 7 (36.8%) out of 19 people whose oxalate excretion was more than 40 mg/24 h had this type of stone. Also, 12 (63%) out of 19 people whose oxalate excretion was above 40mg/24 h had no stones. However, in 75.34% (55.73) of people who had CaOx kidney stones, the excretion of oxalate was within normal range, which was 10 - 40 mg/24 h.
| Variables | Kidney Stone | P-Values | ||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Age groups | 0.004 | |||
| 20 > | 26 (35.61) | 7 (13.46) | 33 (26.4) | |
| 20 - 35 | 12 (16.43) | 8 (15.38) | 20 (16.0) | |
| > 35 | 35 (47.94) | 37 (71.15) | 72 (57.6) | |
| Gender | 0.177 | |||
| Men | 27 (36.98) | 13 (25) | 40 (32.0) | |
| Female | 46 (63.01) | 39 (75) | 85 (68.0) | |
| Oxalatemg/24 h | 0.003 | |||
| 10 > | 11 (15.06) | 3 (5.76) | 14 (11.2%) | |
| 10 - 25 | 41 (56.16) | 21 (40.38) | 62 (49.6) | |
| 25 - 40 | 14 (19.17) | 16 (30.76) | 30 (24.0) | |
| > 40 | 7 (9.58) | 12 (23.07) | 19 (15.2) | |
aValues are expressed as No. (%) unless otherwise indicated.
4.2. Conventional PCR Results with FRC.F/FRC.R Primer
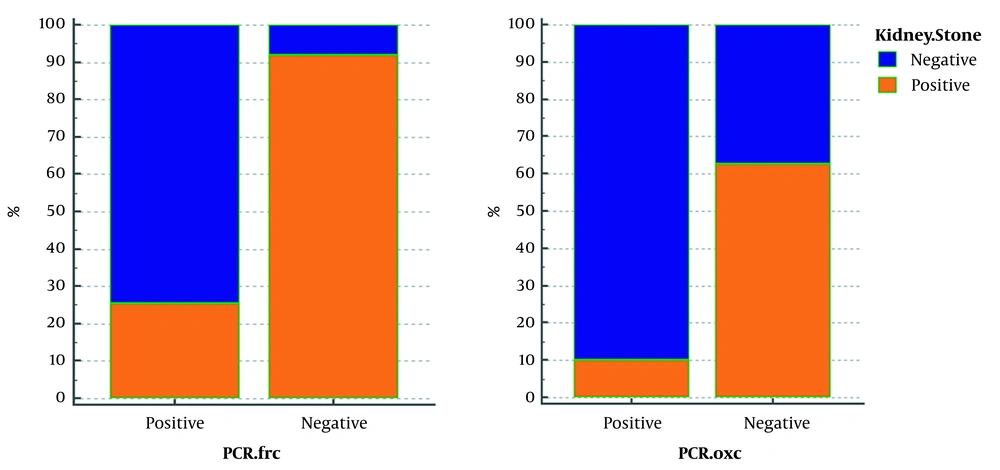
The FRC gene was amplified to identify Oxalobacter formation with specific primers of this gene. Of 52 samples without CaOx kidney stones, 47 samples were reported positive by PCR reaction with FRC gene primers. The 316-bp band confirmed the presence of this gene out of 73 samples with oxalate kidney stones. Seventy-two samples were reported negative by PCR assay, using OXC gene primers, indicating no band, while in 9 of 52 samples, the observation of the 413bp band showed the presence of this gene in people without kidney stones. According to Table 2, in 125 patients studied, 90.38% who did not have CaOx stones (n = 52) had a positive PCR result, and in 78.08% who had this type of kidney stones (n = 73), PCR results were negative for FRC gene. In fact, 91.93% of the patients with negative (n = 62) FRC had stones, and 74.63% of people who were positive for this gene (n = 63) did not have stones. PCR results for 73 patients with oxalate kidney stones on the OXC gene show that 98.6% of these patients had negative Oxalobacter, but this percentage was reported to be similarly high in those without stones (82.69%). Furthermore, PCR results of this gene were positive only in 17.3% of people who did not have stones. As shown in Figure 1, in fact, 62.06% of people whose OXC gene was negative (115/125) had stones, and 90% of people whose PCR was positive for this gene (10/125) had no stones.
| Variables | Kidney Stone | P-Values | ||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| FRC gene (PCR) | 0.0001 | |||
| Positive | 16 (21.91) | 47 (90.38) | 63 (50.4) | |
| Negative | 57 (78.08) | 5 (9.61) | 62 (49.6) | |
| OXC gene (PCR) | 0.0013 | |||
| Positive | 1 (1.36) | 9 (17.30) | 10 (8) | |
| Negative | 72 (98.63) | 43 (82.69) | 115 (92) | |
| FRC gene (qPCR) | 0.0001 | |||
| Positive | 2 (2.73) | 32 (61.53) | 34 (27.2) | |
| Negative | 71 (97.26) | 20 (38.48) | 91 (72.8) | |
| OXC gene (qPCR) | 0.0001 | |||
| Positive | 3 (4.1) | 21 (40.38) | 25 (20) | |
| Negative | 70 (95.89) | 31 (59.61) | 100 (80) | |
a Values are expressed as No. (%) unless otherwise indicated.
4.3. The Results of Sensitivity and Specificity of FRC and OXC Genes Using PCR Techniques
With this method, in 125 samples, 62 cases were negative for FRC gene, and through OXC gene we were able to identify only 10 positive samples. The study of the relationship between sensitivity and specificity of FRC and OXC genes showed that the positive predictive value (PPV) of OXC gene is 100%, and the negative predictive value (NPV) of this gene is 53.91% (Table 3).
| Comparison of Gene Efficiency | FRC | Total, No. (%) | Predictive Value | |
|---|---|---|---|---|
| Positive | Negative | |||
| OXC | ||||
| Positive | True positive: 10 | False positive: 0 | 10 (8.0) | PPV: 100.000% |
| Negative | False negative: 53 | True negative: 62 | 115 (92.0) | NPV: 53.913% |
| Total | 63 (50.4%) | 62 (49.6%) | 125 (100) | |
| Accuracy OXC 57.60% | ||||
| 48.44% to 66.39% (95% CI) | Sensitivity: 15.873% | Specificity: 100.000% | Accuracy FRC | |
4.4. Real-time PCR Analysis (qPCR)
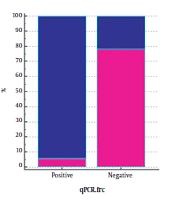
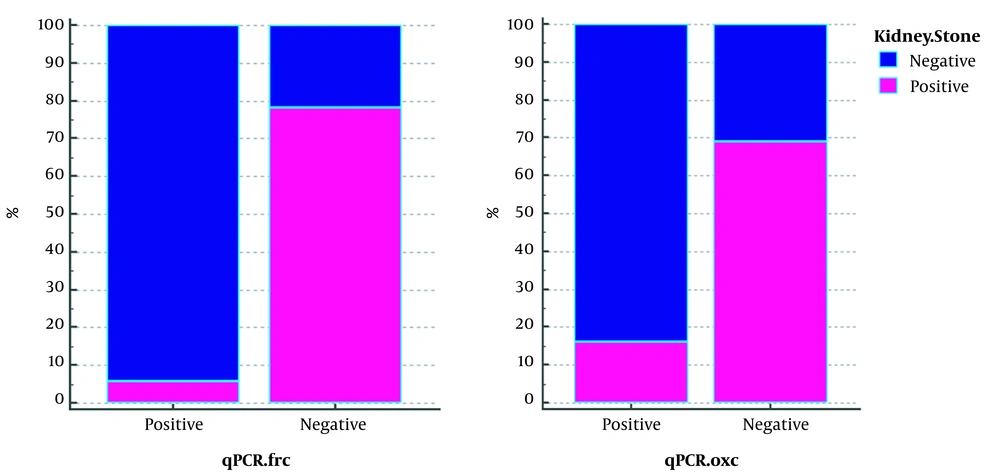
The results of qPCR of FRC gene were negative in 71 (97.26%) of 73 patient who had stones and positive in 32 (61.53%) of 52 people without stones (Table 4). The results showed that 94.11% of qPCR-positive (34.125) samples for FRC gene without stones and 78.02% of negative qPCR samples (91.125) for this gene had stones. The outcome of qPCR of OXC gene was negative in 70 (95.89%) out of 73 patients who had stones, but the results were negative in 31 people (59.61%) out of 52 people who did not have stones. We also found that 84% of the 25 qPCR-positive samples for this gene had no stones, and 69% of the 100 qPCR-negative samples for this gene had stones (Figure 2).
| Comparison of Gene Efficiency | FRC | Total, No. (%) | Predictive Value | |
|---|---|---|---|---|
| Positive | Negative | |||
| OXC | ||||
| Positive | True positive: 20 | False positive: 4 | 24 (29.2) | PPV: 83.33% |
| Negative | False negative: 14 | True negative: 87 | 101 (80.8) | NPV: 86.14% |
| Total | 34 (27.2) | 91 (72.8) | 125 | |
| Accuracy OXC 85.60% | ||||
| 78.20% to 91.24% (95% CI) | Sensitivity: 58.824% | Specificity: 95.6% | Accuracy FRC | |
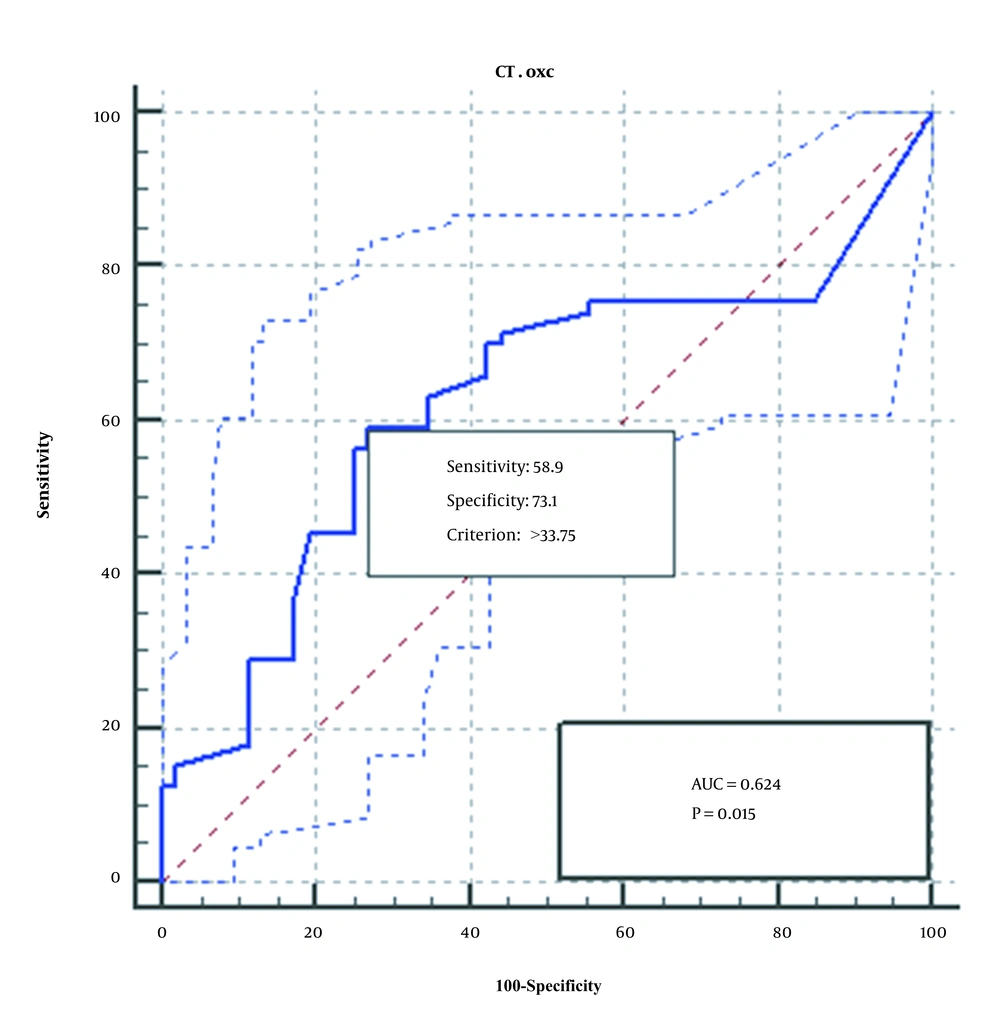
4.5. ROC Test Results
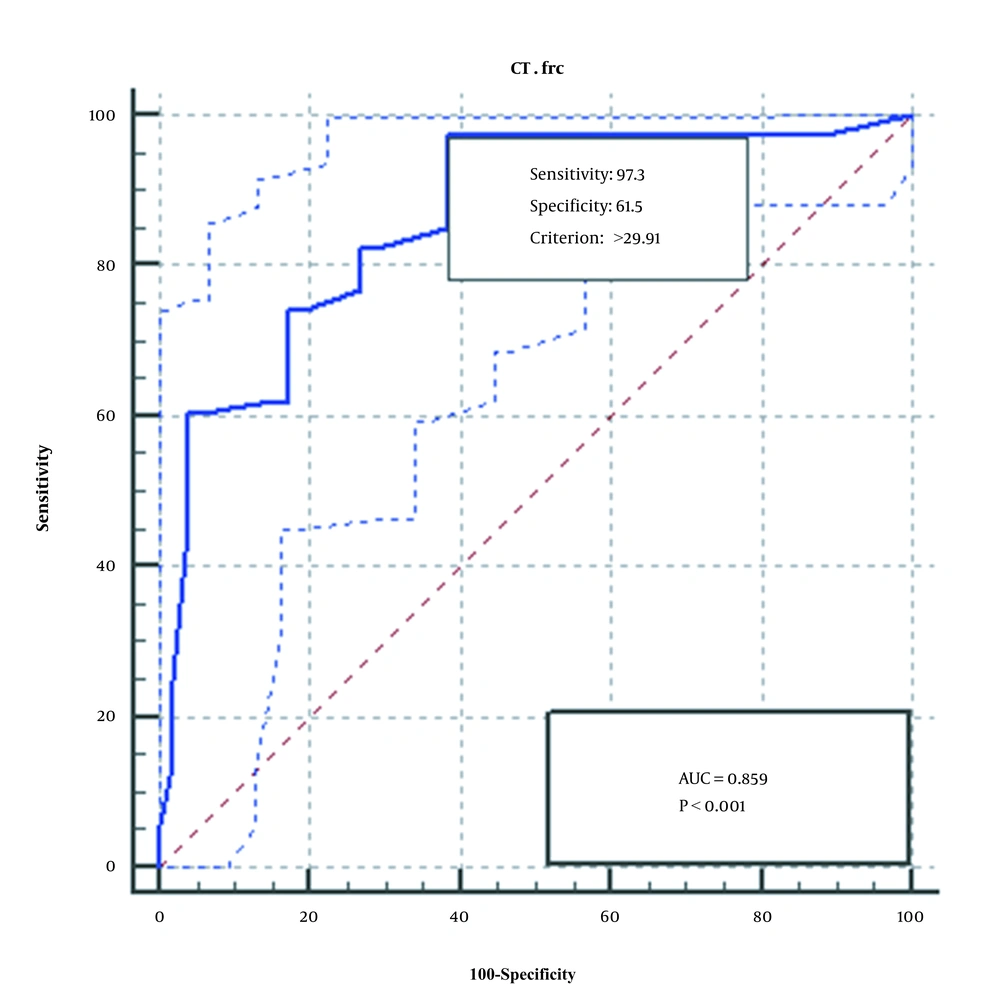
The ROC diagram obtained from the analysis of the qPCR method of the two genes, FRC and OXC, is shown in Figures 3 and 4. As seen, qPCR method for FRC gene is more sensitive, specific, and accurate than OXC gene.
4.6. Sensitivity and Specificity of FRC and OXC Genes Using qPCR Techniques
In the launched qPCR method, the sensitivity and specificity of OXC gene compared with FRC gene were reported to be 58.82% and 95.60%, respectively. Compared to PCR method, sensitivity increased significantly, while specificity decreased slightly.
4.7. Sensitivity of the Studied Methods
PCR technique could identify more positive samples compared to qPCR technique on FRC gene. The qPCR method on the FRC gene had a sensitivity of 49.2% and a specificity of 95.16% compared to the PCR method for this gene (Table 5). The qPCR method on the OXC gene had a sensitivity of 50% and a specificity of 83.4% compared to the PCR method for this gene (Table 6).
| Comparison of Gene Efficiency | PCR | Total, No. (%) | Predictive Value | |
|---|---|---|---|---|
| Positive | Negative | |||
| qPCR | ||||
| Positive | True positive: 31 | False positive: 3 | 34 (27.2) | PPV: 91.18% |
| Negative | False negative: 32 | True negative: 59 | 91 (72.8) | NPV: 64.84% |
| Total | 63 (50.4) | 62 (49.6) | 125 (100) | |
| Accuracy OXC | Sensitivity: 49.206% | Specificity: 95.161% | Accuracy FRC | |
| Comparison of Gene Efficiency | PCR | Total | Predictive Value | |
|---|---|---|---|---|
| Positive | Negative | |||
| qPCR | ||||
| Positive | True positive: 5 | False positive: 19 | 24 (19.2) | PPV: 20.83% |
| Negative | False negative: 5 | True negative: 96 | 101 (80.8) | NPV: 95.05% |
| Total | 10 (8) | 115 (92) | 125 (100) | |
| Accuracy OXC 80.80% | ||||
| 72.79% to 87.29% (95% CI) | Sensitivity: 50.000% | Specificity: 83.48% | Accuracy FRC | |
4.8. Cohen's Kappa Coefficient Between PCR and qPCR Techniques
In this study, the kappa coefficient for measuring the relationship between PCR and qPCR results of FRC gene was 0.44 (with a confidence interval of 0.3 to 0.58 and a standard error rate of 0.07), indicating agreement or moderate correlation between the two techniques for the FRC gene. Kappa coefficient for measuring the relationship between PCR and qPCR results of OXC gene was 0.204 (with a confidence interval of - 0.4 to - 0.001 and a standard error rate of 0.1), which indicates a relatively weak agreement or relationship between the two techniques for the OXC gene (Table 7).
| Value of K | Strength of Agreement |
|---|---|
| < 0.20 | Poor |
| 0.21 - 0.40 | Fair |
| 0.41 - 0.60 | Moderate |
| 0.61 - 0.80 | Good |
| 0.81 – 1.00 | Very good |
5. Discussion
Kidney stone is an increasingly pervasive disease that affects almost 1 - 19.1% of the population (1, 2). Previous studies investigating the bacterial contribution to the disease have focused on intestinal bacteria that degrade oxalate, in particular O. formigenes (16, 17).
Several recent studies have evaluated the normal intestinal microflora in kidney stone disease and described it as "an imbalance of the natural microflora" but there was no consensus among findings (16, 18-21). Accordingly, the manner by which normal intestinal microflora changes, the probable role of O. formigenes colonization as a stimulus, and the normal microflora of the urinary tract and intestines, leading to nephrolithiasis, are still not clear (16, 22). The aim of this study was to determine the effect of natural intestinal microflora, including O. formigenes in kidney stone disease. Kidney stones are believed to be a multifactorial disease that affects lifestyle as well as eating habits. In the general population, CaOx kidney stones are the most common, especially in men, with the third decade of life that is the average age of the onset of the symptoms (9, 16, 22).
In the present study, 63.01% of all people with stones (73 people) are women, but since the ratio of women to men population is approximately 2:1, this disproportion can affect results. In other words, it can be said that the frequency of urinary stones among men (40 people) is 67.5%, and among women (85 people) it is 46%, which shows that the incidence of this stone is higher in men (14, 19, 23). In the present study, the incidence of CaOx stone in young people (< 20) with a frequency of 78.78% is higher than older ones. In addition, a significant relationship was observed between age and the incidence of CaOx stone (P = 0.004); the lower average age of people with this stone is the evidence of this claim (29/27 years). Urinary oxalate levels are a major risk factor for the formation of CaOx kidney stones (23). It has been long hypothesized that colonization of the gut by bacteria with oxalate degradation capacity is inversely related to the risk of kidney stones. However, there are contradictions in different studies, which affect the type of food consumed and digestion (19). It demands to cultivate and culture O. formigenes, especially that it is present in tiny amounts in feces, which requires special anaerobic conditions (24). Therefore, in almost all studies with O. formigenes, conventional PCR was used to identify gastrointestinal bacteria (24-26), but in one study, a real-time PCR was used to count the number of O. formigenes in a healthy group (5).
In one study, 45.6% of patients with CaOx stone had O. formigenes colonization. In another study, colonization in 33% of patients with bladder inflammation and 65% of patients in the control group was detected (25-27). Our study, based on FRC gene and PCR technique, showed that O. formigenes colonization is 90.38 and 21.91% in healthy individuals and people with kidney stones, respectively. Also, the study of this gene with qPCR technique revealed the colonization of this bacterium in healthy individuals at 6.53% and in individuals with stones at 2.73%. However, we found different results based on OXC gene. According to the PCR technique, O. formigenes colonizations are 17.30 and 1.36% in healthy individuals and those with kidney stones, respectively. The qPCR technique shows that colonization of this bacterium in healthy individuals and others with kidney stones is 40.38% and 4.1%, respectively. The reason for the absence of O. formigenes colonization in 30 - 40% of healthy individuals and 70-60% of urinary patients is not yet clear. However, the use of antibiotics is thought to be a factor in colonization (28, 29). In the study of Batislam et al. for the first time, a qPCR method based on the OXC gene was used to identify the exact extent of colonization in patients with kidney stones. This study concluded that the number of O. formigenes is significantly lower in patients with metabolic disorders than in ones without metabolic disorders (30).
In the present study, the comparison of PCR results of OXC and FRC genes shows that FRC gene is more efficient in isolating Oxalobacter. Although the specificity of qPCR reaction has increased compared to conventional PCR on OXC gene, OXC gene is not a suitable diagnostic indicator in PCR and qPCR technique; however, it can play a helpful role. Finally, by examining the results of the number of positive diagnostic cases by FRC and OXC gene regions and using two conventional PCR and qPCR techniques, it was found that FRC gene is more sensitive than OXC gene, and OXC gene is more specific (31, 32).
In the present study in Qom province, O. formigenes was detected more in the control group than the patients with urinary stones, which is consistent with other studies. In the PCR technique, due to the specificity of 100% of OXC gene and its low sensitivity compared to FRC (15.87%), we can trust its positive answers with full confidence based on its PPV, while 53.91% of the negative answers can be trusted (31, 32). Investigation of the relationship between sensitivity and specificity of FRC and OXC genes by qPCR method showed that the PPV of OXC gene is 83.33% and the NPV of this gene is 86.14% qPCR technique in comparison with PCR technique on FRC gene showed NPV and PPV of this gene 64.84% and 91.18% and on OXC gene 95.5% and 20.83%, respectively. Although understanding the role of O. formigenes in oxalate metabolism is still limited, it is hypothesized that the absence of this bacterium may be associated with higher intestinal oxalate uptake (31).
Degradation of oxalate by O. formigenes is expected to reduce urinary oxalate excretion, and several studies have linked the absence of O. formigenes to higher urinary oxalate excretion (25, 33). Nevertheless, other studies have revealed significant differences in oxalate excretion urine among patients who did not show a positive or negative test for O. formigenes (34).
In our study, there was no significant difference in the mean oxalate uptake between 73 patients with CaOx stone and 52 control patients without this type of stone. Also, according to Table 2, most of the people with stones whose PCR result was positive (62.5%) or negative (54.38%) for FRC gene have oxalate excretion in the normal range of 25 - 10 mg / 24h. Also, 56.94% of patients with kidney stones whose PCR test was negative for OXC gene had oxalate excretion in the normal range of 25 - 10 mg / 24h. This is true of the results provided by the qPCR method for both genes. Knight et al. reported in 38 CaOx rock manufacturers that uptake was not significantly different in those with and without bacteria (34). In addition, a new study showed that 24-hour urinary oxalate and plasma oxalate excretion rates were significantly lower in patients with O. formigenes cloned compared with O. formigenes-negative patients in a standard diet (34, 35).
The relationship between recurrent CaOx stone disease and O. formigenes deficiency was evaluated in a study of 247 patients with calcium oxalate stone formation and 259 control groups (9). The Odds ratio for the formation of a recurrent stone during colonization was 0.3, indicating a 70% reduction in stone risk. Surprisingly, there was no difference in urinary oxalate excretion between people whose colons’ bacteria had been colonized and those whose intestines’ bacteria had not been colonized. Both may be highly variable due to the results of oxalate excretion, despite sufficient sample size, especially that oxalate levels and dietary calcium were not controlled (9).
The limitations of this study include the relatively small number of people to deal with the variables under study. Also, normal distribution was not observed in the study population. In addition, dietary oxalate may have a significant effect on the risk of developing CaOx kidney stones. In this study, individual dietary information was not generally available, so differences in oxalate levels could not be assessed. As a result, an accurate quantification tool such as qPCR is needed to measure these responses and the variables under consideration.