1. Background
Hemodialysis is one of the renal replacement therapies in patients with end-stage renal failure. Hemodialysis is the 5th most common procedure for patients aged 45 - 64 years (1, 2). Due to the fact that the procedure of hemodialysis requires prolonged vascular access, patients undergoing chronic hemodialysis are at increased risk of blood-transmitted infections (3). The risk of infection is related to several factors including environmental factors such as contaminated devices, equipment and supplies, environmental surfaces, and personnel’s hands (4, 5), as well as patient related factors such as the number of years spent on dialysis therapy (dialytic age), partial immunosuppressant, regular hospitalizations, and parenteral interventions frequency (6-8).
Hepatitis C virus (HCV) infection is a serious public health problem (9) and the most significant cause of liver disease in patient receiving dialysis for a long term (10). HCV still remains frequent in patients undergoing maintenance hemodialysis and causes substantial morbidity and mortality in them (11, 12). The world health organization (WHO) estimates the universal prevalence of HCV infection around 3% (13, 14). Since the prevalence of HCV in patients undergoing hemodialysis is considerably higher than in general population, it is a permanent concern for hemodialysis units (15, 16). Frequency of blood transfusions and HCV prevalence in the blood units, as well as HCV prevalence in the patients undergoing hemodialysis simultaneously in the same environment are associated with an increased risk for nosocomial transmission of HCV infections (17). An early and accurate diagnosis of HCV in patients with end-stage renal disease should be performed to control the HCV transmission in hemodialysis units (18, 19).
Patients with chronic renal diseases undergoing hemodialysis are recommended to be screened for HCV infection (19). Currently, the HCV diagnosis is made by 2 main methods; detection of anti-HCV using enzyme-linked immunosorbent assay (EISA), which determines a present or resolved past infection, and detection of HCV-RNA in serum via PCR method, which indicates an active infection (20-23). Detecting reactive antibody against HCV using ELISA is the commom screening test for HCV infection, but it has limitations. The prolonged window period (up to 18 months) and delayed production of anti-HCV in such patients under immunosuppression result in false negatives (up to 22%) (23). In addition, several studies reported different results about ELISAs in screening HCV infection. In contrast, HCV RNA does persist in serum and is easily detectable in patients with impaired immune functions such as the ones underwent chronic hemodialysis within the first 2 weeks of infection (24).
The current study aimed at evaluating the prevalence of HCV infection in a hemodialysis population, and comparing serological (ELISA) and molecular (PCR) methods to detect HCV infection in Sari, Iran (Mazandaran province) from January to July 2015.
2. Methods
The current cross sectional study was conducted from January to July 2015; serum samples of 162 patients undergoing chronic hemodialysis were collected in 2 hemodialysis units in Sari, Iran. The local ethical committees approved the study. The demographic data and patient characteristics such as gender and age, in addition to other data including the cause of renal disease, duration of hemodialysis as well as history of blood transfusion, previous kidney transplantation, drug use, liver disease, and hepatitis B virus (HBV) infection were collected in a datasheet. To perform anti-HCV test, HCV-PCR and biochemical enzymes (blood urea nitrogen, creatinine, alanine transaminase, aspartate aminotransferase, and alkaline phosphatase), 10-mL blood samples were collected from all patients, serum and plasma were separated by centrifugation and stored at -70°C.
RNA Extraction: RNAs were extracted from plasma samples by RIBO-prep nucleic acid extraction kit (AmpliSens®, Russia). HCV RNA was isolated from samples by the following procedures: A100 µL of prepared samples was added to the tubes with 300 µL solution and vortexed thoroughly; then, tubes were centrifuged for 5 seconds to make sure that there were no drops on the cap, and incubated at 65°C for 5 minutes. A 400 µL of solution was added for precipitation and votrexed; then, centrifuged for 5 minutes at 13,000 revolutions per minute (rpm). Next, the supernatant was carefully removed without disturbing the pellet using a vacuum aspirator and new tips, then washed with 500 µL of washing solution 3 and centrifuged at 13,000 rpm. The supernatant was carefully removed and 200 µL of washing solution 4 was added to each tube and centrifuged at 13,000 rpm; then, the supernatant was carefully removed. Finally, all tubes were incubated with open caps for 5 minutes, dried at 65°C, and 50 µL of sterile distilled water was added into each tube, and vortexed. Tubes were incubated at 65°C for 5 minutes and centrifuged at 13,000 rpm, then RNA quantification was determined using a spectrophotometer and the resulted residue was solved for next stages.
2.1. Real-Time PCR
Total RNA was isolated from samples and real time PCR was performed using HCV-FRT PCR Kit (AmpliSense, Russia) according to the manufacturer’s instructions. Hepatitis C virus detection from plasma samples along with the internal control sample (IC) was performed under isolation conditions to control the accuracy of results. Briefly, 15 µL of the master mix was added to the tubes containing 10 µL of extracted RNA. AmpliSens® HCV-FRT PCR kit contained hotstart, which greatly reduced the frequency of nonspecifically primed reactions. The hotstart was guaranteed by separation of nucleotides and Taq polymerase by a chemically modified polymerase (TaqF). The latter was activated by heating at 95°C for 15 minutes. The IC amplification product was detected in the FAM channel. The HCV cDNA amplification product was detected in the JOE/HEX channel. The positive control of extraction, positive control-1-HCV, was detected in FAM (IC) and JOE/HEX (HCV) channels. The positive control of amplification, PIC2 HCV (C+), was a complex control for HCV and IC. It is detected in FAM (IC) and JOE/HEX (HCV) channels. The PCR program is presented in Table 1. PCR reaction was performed using a Rotor Gene 6,000 real-time PCR Machine (Corbett Research, Australia). After this stage, using a computer program, results were immediately determined in programming RT-PCR; besides, positively of test, amount and quantity of virus was determined.
| Step | Temperature, °C | Time | Fluorescence Detection | No. of Cycles |
|---|---|---|---|---|
| 1 | 50 | 15 min | - | 1 |
| 2 | 95 | 15 min | - | 1 |
| 3 | 95 | 5 s | - | 5 |
| 60 | 20 s | - | ||
| 72 | 15 s | - | ||
| 4 | 95 | 5 s | - | 40 |
| 60 | 20 s | FAM and JOE | ||
| 72 | 15 s | - |
2.2. Anti-HCV Test
Anti-HCV was tested using 500 µL serum sample, HCV antibody ELISA kit (DSL, Webster, TX, USA) and fully automated ELISA plate reader (Dynex, USA).
2.3. Biochemical Assessments
Venous blood samples were collected and serum was separated by centrifugation. The serum value of blood urea nitrogen (BUN), creatinine (Cr) and liver enzymes (aspartate aminotransferase [AST], alanine transaminase [ALT] and alkaline phosphatase [ALP]) were measured enzymatically (Pars Azemoon co.) using Auto Analyzer HITACHI 902.
3. Results
A total of 162 patients with the end-stage renal failure undergoing chronic hemodialysis, with the mean ± SD age of 68.81 ± 13.76 years (range, 30 to 94) were included in the current study; in which 98 (60.5%) patients were male and 64 (39.5%) female. The average length of time spent on dialysis therapy was 35.67 ± 29.99 (mean ± SD) months (Table 2). Among them, 117 (72.2%) patients underwent hemodialysis 3 times a week, 38 (23.5%) twice a week, and 7 (4.3%) once a week; 69 (42.6%) patients had blood transfusion history and 93 (57.4%) did not (Table 2). Transfusion frequency is presented in Table 3. Moreover, 6 patients (3.7%) had kidney transplantation previously.
| Characteristics | Data |
|---|---|
| Age, y | 68.81 ± 13.76 |
| Range | 30 - 94 |
| Gender, male/female | 98/64 (60.5/39.5) |
| Duration of hemodialysis, months | 35.67 ± 29.99 |
| History of transfusion | 69 (42.6) |
| History of renal transplant | 6 (3.7) |
| History of hepatitis B infection | 3 (1.9) |
| HBS Ag positivity | 5 (3.1) |
aValues are expressed as mean ± SD or No. (%).
| No. of Transfusion | Patients |
|---|---|
| Without transfusion | 93 (57.4) |
| 1 | 22 (13.6) |
| 2 | 24 (14.8) |
| 3 | 13 (8) |
| 4 | 5 (3.1) |
| 5 | 2 (1.2) |
| 6 | 1 (0.6) |
| 11 | 1 (0.6) |
| Undefined | 1 (0.6) |
| Total | 162 (100) |
aValues are expressed as No. (%).
Interviews revealed that 3 patients (1.9%) had the history of HBV infection. However, none of the patients had the familial history of hepatitis. HBs Ag assessments illustrated that 5 patients (3.1%) were HBS-Ag positive.
Of all the patients, 7 (4.3%) were HCV-ab positive and 155 (95.7%) were HCV-ab negative. Additionally, 11 patients (6.8%) were HCV-PCR positive, while 151 (93.2%) were HCV-PCR negative. Among 11 HCV-PCR positive patients, 7 (63.6%) were HCV-ab positive and 4 (36.4%) were HCV-ab negative. Furthermore, HCV-ab test was not positive in any of the HCV-PCR negative patients. Comparison between the results of the 2 methods (HCV-an and HCV real-time PCR) is presented in Table 4.
| HCV-PCR | |||
|---|---|---|---|
| Positive | Negative | Total | |
| HCV-ab | |||
| Positive | 7 (63.6) | 0 | 7 (4.3) |
| Negative | 4 (36.4) | 151 (100) | 155 (95.7) |
| Total | 11 (100) | 151 (100) | 162 (100) |
aValues are expressed as No. (%).
Among HCV-PCR positive patients, 7 (63.6%) were male and 4 (36.4%) female. In other words, 7.1% and 6.2% were HCV-PCR positive among males and females, respectively, but the difference was statistically insignificant (P = 0.825).
None of the patients with the history of kidney transplantation showed HCV-PCR positivity. In contrast, 7.1% of the patients with no history of kidney transplantation were HCV-PCR negative. However, the difference between the 2 groups was statistically insignificant (P = 0.500).
Among HCV-PCR positive patients, 5 (45.5%) had the history of blood transfusion. In other words, among patients who received previous transfusion 7% and among the ones with no transfusion history 6.6% were HCV-PCR positive. The difference was statistically insignificant (P = 0.910).
Interviews revealed that 3 (1.9%) patients had the history of HBV infection and none of the patients had the familial history of hepatitis. HBs Ag assessments illustrated that 5 (3.1%) patients were HBs Ag positive. None of HCV-PCR positive patients had HBV infection history. Among HBs Ag negative patients, 6.9% were HCV-PCR positive. The difference between the 2 groups was insignificant (P = 0.637).
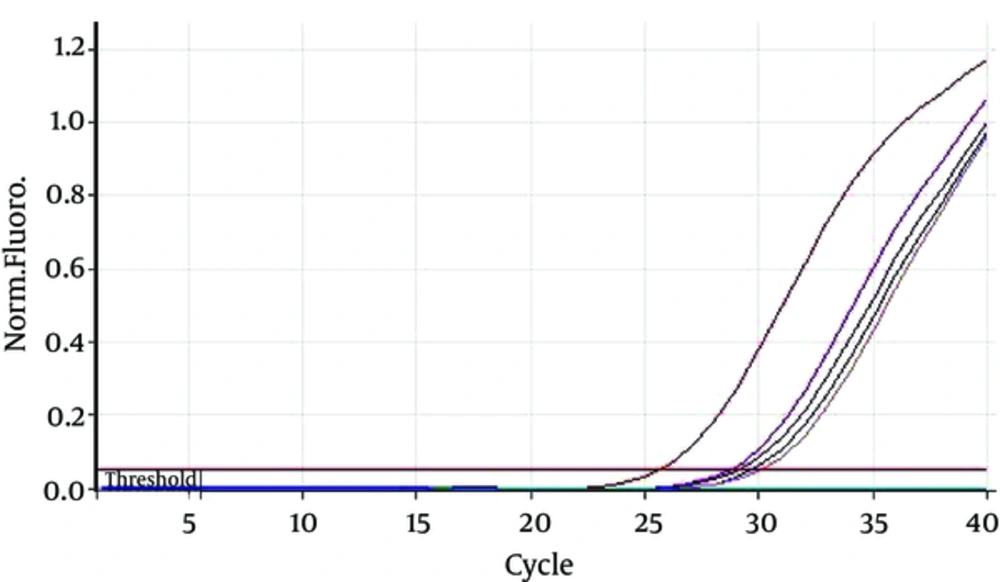
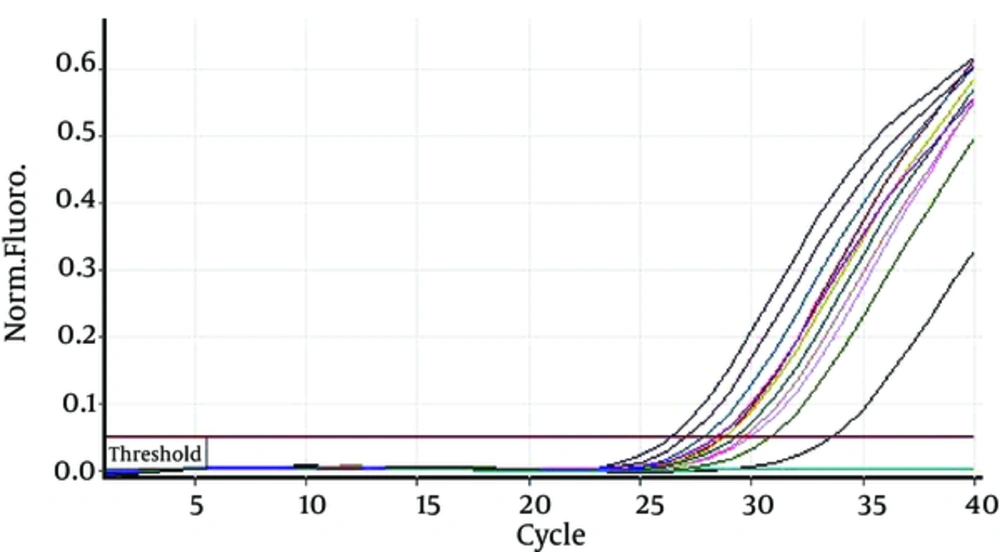
Duration of hemodialysis in HCV-PCR positive patients was 65.55 ± 55.10 months and in HCV-PCR negative cases was 33.49 ± 26.42; there was a significant difference between HCV-PCR positive and negative patients (P = 0.001). Furthermore, HCV-PCR positive cases underwent hemodialysis 2.82 ± 0.41 hours per week and HCV-PCR negative patients underwent hemodialysis 2.67 ± 0.56 hours per week; the difference was statistically insignificant (P = 0.387) (Figures 1 and 2).
Biochemical parameters are represented in Table 5. AST values were significantly different between 2 groups (P = 0.128). Although the values of Cr, ALT and ALP were higher in HCV-PCR positive cases, the difference was statistically insignificant (P > 0.05).
| Biochemical Parameters | HCV-PCR Positive | HCV-PCR Negative | Total | P Value |
|---|---|---|---|---|
| BUN | 83.18 ± 47.59 | 97.96 ± 50.32 | 96.93 ± 50.13 | 0.342 |
| Cr | 8.09 ± 2.09 | 6.99 ± 2.89 | 7.06 ± 2.85 | 0.128 |
| AST | 43.45 ± 84.01 | 16.51 ± 8.83 | 18.47 ± 24.33 | < 0.001 |
| ALT | 19.90 ± 16.46 | 15.24 ± 11.20 | 15.55 ± 11.61 | 0.400 |
| ALP | 416 ± 348.88 | 341.80 ± 278.62 | 347.47 ± 283.83 | 0.506 |
4. Discussion
Since the prevalence of HCV in patients undergoing hemodialysis is considerably higher than in general population (25), chronic renal patients undergoing hemodialysis are recommended to be screened for HCV infection (19). Anti-HCV tests are common screening methods used for HCV infection, but they may not accurately reflect true HCV status (26). Despite the advantages of tests in antibody detection such as easy application, relatively low cost, and enhanced sensitivity of recent generations, they still have limitations. The window period between acute infection and antibody production may be more prolonged in such patients with immunodepression (27, 28). Due to the slow or delayed seroconversion in the immunodeficiency state; an interval of up to 18 months is reported in patients undergoing hemodialysis (29). Furthermore, another weakness of anti-HCV tests is their incapability to differentiate the present active infection from the recovered past infection (30). In contrast, direct viral detection such as HCV-PCR testing is an effective method; since HCV RNA is detectable in serum within 2 weeks of infection; it vanishes if the infection recovers and persists in chronic infections (31, 32). Therefore, HCV-PCR testing can distinguish recovered past infections from present active infections and detect HCV RNA before preseroconversion and antibody production (33, 34). Accordingly, PCR method is recommended in addition to HCV antibody testing in this population (35). HCV-PCR testing has its own limitations for mass screening, including the cost and high technical skill requirements (36).
HCV-ab was positive in 4.3% of cases, but HCV-RNA using PCR was positive in 6.8%. The current study results showed lower HCV prevalence than other studies such as those of Caramelo et al. (37), Jadoul et al. (38), makhlough et al. (22), and Moini et al. (36). The decreased prevalence of HCV in the current study in comparison to other studies may be due to multiple factors including HCV screening of blood units, erythropoietin prescription to treat anemia in patients undergoing hemodialysis, using dedicated hemodialysis machine, hygiene improvement in hemodialysis units such as dialysis of HCV positive patients in separate wards, and HCV knowledge improvement in medical professionals.
In the present study, all HCV-ab positive samples showed HCV RNA positivity using PCR method; thus, no false positive result was observed in HCV-ab test; however, 36.4% false negative results were observed. Therefore, in comparison with HCV-PCR, HCV-ab test has the sensitivity of 63.6% and specificity of 100%. According to the current study statistical analysis, the specificity of HCV-PCR test was significantly higher than that of HCV-ab test (P < 0.0001). The current study results were in accordance with those of Caramelo et al. (37). Hence, in patients undergoing hemodialysis with no detectable amounts of HCV antibody in their circulation, HCV-PCR is recommended.
In the current study, hemodialysis period was significantly longer in HCV positive than HCV negative patients. The findings were in agreement with those of Makhlough et al. (22) and Duong et al. (39). It seems that the risk of HCV infection is increased with the increase of hemodialysis period; therefore, these patients require more serious cautions.
The current study findings showed that HCV prevalence was insignificantly different in patients with blood transfusion or kidney transplantation history and patients without such history. It was in contrast with the findings of Duong et al. (39) and Tu et al. (40). At present, nosocomial transmission within the dialysis centers is the main cause of HCV transmission.
In the present study, 1.9% of cases had the history of HBV infection and 3.1% were HBs Ag positive; and none of HCV-PCR positive patients had HBV infection history. The current study results were in contrast with those of Tu et al. (40), which demonstrated significant relationship between HCV and HBV infection in patients undergoing hemodialysis. The increasing use of HBV vaccine and screening the blood units result in remarked decrease in HBV infection. HCV still remains frequent in patients undergoing maintenance hemodialysis, due to the lack of HBV vaccine.
In the present study, the values of Cr, ALT, and ALP were insignificantly different in HCV positive and negative cases (P > 0.05); but AST values were significantly different between the 2 groups (P = 0.128). The current study results were in contrast with those of Caramelo et al. (37) and Duong et al. (39), which illustrated that liver enzymes were significantly higher in HCV positive cases. Most of the individuals newly infected with HCV (60% - 70%) are asymptomatic (41). Later, HCV infection tends to become a chronic infection in approximately 75% of cases (42); subsequently, chronic HCV infection leads to progressive liver diseases such as end-stage liver disease, cirrhosis or liver cancer in 15% - 25% of patients; but they most often have an indolent coarse. Moreover, liver enzyme values have lower specificity in patients undergoing hemodialysis than normal individuals; and serum values of liver enzymes were not directly correlated with the degree of liver damage and HCV titer (43, 44).
4.1. Conclusions
The current study findings demonstrated that the specificity of of HCV-RNA detection test was significantly higher than that of conventional HCV-ab test. As patients undergoing hemodialysis did not have detectable antibody in their serum, using PCR method is highly remarkable.