1. Context
Papillomavirus is a large family of viruses belonging to the family Papovaviridae, the genus Papillomavirus. To date, more than 200 types of human papillomavirus (HPV) have been recognized based on DNA sequence (1). About 40 types of HPV are sexually transmitted and affect fully differentiated squamous epithelium of the skin and mucosal epithelium of anogenital and non-genital sites, among which 15 types can cause cervical cancer (CC) (2). The immune system spontaneously clears most HPV infections (approximately 90%) within 1 or 2 years. ‘High-risk’ mucosal HPV types, predominantly types 16, 18, 31, 33, and 35, have been associated with most cervical, penile, vulvar, vaginal, anal, and oropharyngeal cancers and pre-cancers (3). In general, HPV is thought to be responsible for more than 90% of anal and cervical cancers, about 70% of vaginal and vulvar cancers, and more than 60% of penile cancers (4). In addition, HPV16 and HPV18 are accountable for approximately 70% of CC worldwide (5).
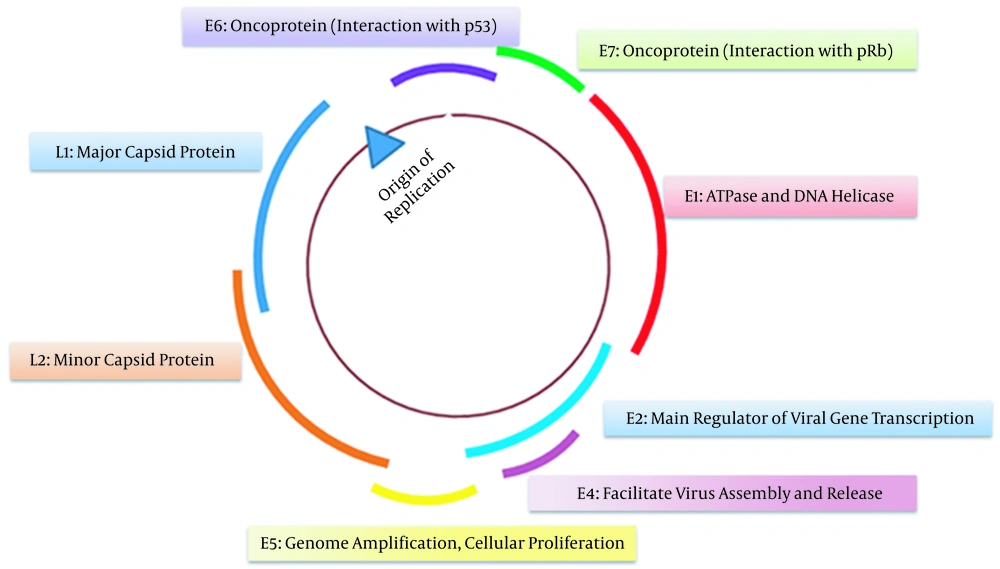
Structurally, HPV16 and HPV18 are small non-enveloped viruses with icosahedral capsid enclosing a circular double-stranded DNA. The total length of viral DNA is about 8000 base pairs (8 kb), encompassing eight open reading frames transcribed as polycistronic messages from a single DNA strand. Based on functional properties and location, the genome can be divided into three regions: an early region (E), a late region (L), and a long control region (LCR). The early region contains six ORFs (E1, E2, E4, E5, E6, and E7), which carry out regulatory functions. The late region encodes L1 and L2, the principal virus capsid protein (Figure 1). The L1 region, with 90% similarity, is the most conserved genome region in HPV and is the prime target for serological assay and vaccine development (6). The extended control region is a noncoding region that holds the origin of replication and several transcription factor binding sites. E1 and E2 proteins are responsible for virus amplification (replication and DNA packaging) and maintenance. Persistent HPV infections are reported to be associated with integrating the HPV genome into the host cell genome. The process of integration frequently occurs within E1 or E2 and leads to their loss. This leads to overexpression of viral oncoproteins E6 and E7 and genetic alterations known as drivers of mutation in HPV carcinogenesis.
The development of CC often takes many years or even decades after persistent infection with HPV. This long time window provides a golden opportunity to elucidate HPV‐host interaction, malignant transformation, and carcinogenesis. HPV genotype, viral load, physical state, integration site, and genomic change at the insertion site are suggested as key features with predictive and prognostic values for clinical management of HPV-related cancers (7).
Nearly all locally advanced HPV-related cancers, regardless of their clinical stage, tumor histology, or tumor size, are treated with a similar paradigm of surgery, external or internal radiation therapy, and systemic or regional chemotherapy (8). However, the standard treatment regimen does not reflect the biologic heterogeneity of HPV-related cancers. Therefore, to improve radiotherapy outcomes, it is necessary to optimize the intensity of radiotherapy based on the individualpatient’s responses. Given the advent of next-generation sequencing (NGS) technology and the implementation of omics approaches (genomics, transcriptomics, and proteomics) in clinical oncology, cancer treatments shift from one-size-fits-all toward more comprehensive, multidisciplinary, precision approaches. However, clinically useful biomarkers to guide radiotherapy optimization are not yet available.
In the present brief review, we aimed to summarize recent research on the prognostic value of the physical status of HPV DNA for the survival rate of patients with HPV-related cancers undergoing radiotherapy.
2. Evidence Acquisition
We performed a systematic search using ISI Web of Science, Scopus, PubMed, and Embase, with a combination of the keywords “human papillomaviruses”, “cervical cancer, “radiotherapy”, “biomarkers”, “viral integration”, and “episome”. All types of studies, including observational or retrospective studies, clinical trials, congress abstracts, and case reports or case series, were investigated to find relevant data with no time limitation.
3. Results
3.1. HPV Physical State
Following infection, the HPV virus can remain in its episomal form or become integrated into the host genome. Besides, both episomal and integrated patterns can be present simultaneously (9). Two main mechanisms of HPV integration into the genomic DNA include looping (which may result in viral-host DNA concatemers) integration and direct insertion. In addition, HPV can be integrated into the host genome as the single-copy or multi-copy of head-to-tail tandem repeats. Single copy HPV integration chiefly occurs somewhere in the E1 and E2 gene region, which normally regulates the expression of the viral oncogenes E6 and E7. This integration results in disruption of the E1 or E2 open reading frame [ORS], leading to deregulation of negative feedback control of the oncogenes E6 and E7. The E6 oncoprotein forms a physical complex with the p53 cellular tumor suppressor, resulting in P53-mediated G1/S checkpoint dysfunction and cell cycle dysregulation (10).
On the other hand, the E7 oncoprotein binds to the retinoblastoma tumor suppressor protein, pRB, releasing the transcription factor E2F from pRb (11). This results in transcriptional activation of E2F-responsive genes participating in cell-cycle control. These events contribute to HPV oncogenesis and progression of low-grade lesions to neoplastic conditions and, ultimately, carcinoma.
In contrast to one copy integration of the HPV genome, when numerous copies are being integrated, only the E1 or E2 of copies surrounding the cellular DNA will be disrupted, while the remaining copies, E1 and E2 ORFs, will stay intact; thus, they can maintain transcriptional control on E6/E7 (12).
Even though HPV oncogenesis is usually regarded as dependent on viral genomic integration, the rate of HPV DNA integration is quite variable according to HPV genotypes. For instance, the integration rate is reported to be almost 100% for HPV-18 positive CC cases but about 80% for HPV-16 cancer cases (13). The importance of integration in the carcinogenesis of CC is further supported by the fact that HPV DNA has been integrated into merely 3% of premalignant lesions (14-16).
Nevertheless, HPV integration is not the only requisite factor in the carcinogenesis of CC as purely episomal forms of the viral genome have also been detected in HPV-related cancers (15, 17-19). According to preliminary studies, the pathogenesis of HPV episome-related cancer, which is associated with numerous copies of episomal HPV, mainly results from overexpression of the viral oncogenes E6 and E7 (19, 20). However, subsequent studies have suggested some roles for genetic or epigenetic changes resulting in dysregulation of E6/E7 gene expression. For instance, the E2-mediated repression of viral oncogenesis decreased through methylation of E2 binding sites in the viral upstream regulatory region (21, 22). This refers to the impact of HPV episome on HPV-related carcinogenesis apart from E2 disruption secondary to integrating the viral genome into the host genome.
In order to better understand the integration mechanism in the carcinogenic process, the physical state (integrated or episomal) of the viral genome should be determined. Of many different techniques being applied, including Southern blot, fluorescent in situ hybridization, and PCR-based approaches like restriction site PCR and the detection of integrated papillomavirus sequences PCR, none was able to differentiate between the two forms of the HPV genome (23, 24). Discrimination between HPV DNA patterns was also not possible when real-time PCR that can measure both E2 and E6 genes was used (25).
In a study by Joo et al. (26), both in situ hybridization [ISH] and PCR were adopted to identify the HPV integration pattern. Locally advanced cervical tumors were categorized into three groups: the episomal group, the single-copy integrated or multi-copy tandem repetition-integrated pattern group, and the undetectable HPV group. According to their results, what happens first is the disruption of the E2 region without being amplified by PCR after the full integration of the virus. Then, following HPV insertion in the form of multi-copy tandem repetition, given that unbroken E2 genes have been repeatedly put into host cells, both E2 and E6 genes can be traced. Due to the multiplicity of E2 and E6 copies in both episomal and mixed patterns, it was concluded that the sole use of PCR was not sufficient to differentiate between different HPV DNA patterns. They evaluated 204 cervical tumors using both PCR and ISH, although there was significant discordance between these methods. Therefore, since ISH can better differentiate between episomal and multi-copy tandem integrated patterns, it can be combined with PCR to improve discrimination (26).
Lately, integration events, like specifying viral integration sites into the human genome and identifying disrupted genes (esp. oncogenes, tumor suppressor genes, and DNA repair genes) can be successfully detected using the NGS technique (27).
Both targeted (the whole exome sequencing of DNA or polyA-specific RNA sequencing) and untargeted, unbiased (the whole-genome sequencing of DNA or total RNA sequencing) methods have been used in available studies. Some other novel targeted approaches, including capture sequencing technologies, have also been introduced, which are based on DNA enrichment before sequencing and allow for deeper analysis of specific regions, including genome-wide promoter assessment (28).
So far, only a few recurrent sites for HPV integration known as genomic “hotspots”, which are strongly associated with regions in host DNA active in transcription and susceptible to damage, have been identified by whole-genome association studies (23). When DNA replication 2stress ensues from HPV integration, structures called stalled replication forks are developed at regions of the host genome, which sometimes show microscopic homology of 1 to 1 base pairs to the viral genome and have high AT bases. HPV integration can also induce regulatory or mutagenic effects on adjacent genes.
Considering the importance of HPV integration into the human genome as an early key stage in malignancy, recognizing the physical state of the HPV genome in low-grade or atypical lesions can have important clinical implications in both identifying prognostic factors for disease progression and selecting more precise therapeutic approaches (29, 30).
3.2. HPV Physical State as a Prognostic Factor for Radiation Response in CC
Cervical malignancy is caused by high-risk HPV types in nearly all cases. When diagnosed at an early stage, CC can be cured using a surgical approach. As an alternative to surgery or complementary treatment, radiotherapy can also be used for some women with early-stage disease. For more advanced cases, however, a combination of both radiotherapy and chemotherapy is often needed. Given that HPV-related tumors are generally well-responsive to radiotherapy, the implementation of a precise radiotherapeutic approach improves both quality and outcome of treatment. However, even in individuals with similar clinical and morphological parameters of their malignant lesion, including anatomic region, histological type, clinical stage, and degree of cancerous cell differentiation, radiosensitivity of tumor cells is quite variable. Therefore, by identifying some unresponsive to conventional radiotherapy, the radiation therapy schedule can be optimized by elucidating the underlying molecular mechanisms. It is believed that radiosensitivity or radioresistance of tumor cells is related to the biological behavior of their genes. Hence, decoding genetic changes within these cells can aid in improving clinical outcomes (31). As the integration of HPV DNA in host somatic cells is a critical step in carcinogenesis, determining the physical status of the HPV genome and the site of integrating or targeting host genes is presumed to modify the disease-free survival rate (32, 33).
In an early study by Unger et al. (34), the ISH approach was applied to determine HPV-18 DNA (episomal, integrated, or mixed) in primary CC tumors and evaluate the correlation of overall disease-free survival rates with the HPV DNA integration rate. According to their results, the integration of HPV DNA in the primary tumor was strongly associated with treatment failure (34). In another complementary study, the same authors evaluated the association between the integration of disrupted E2 and the overall disease-free survival in patients with HPV 16-positive primary CC tumors (32). They thoroughly examined the whole E1/E2 region by amplifying eight separate fragments spanning the region using ISH. Based on their results, the amount of disrupted E2 in sample cells was significantly correlated with reduced disease-free survival compared to other patients with intact E2 (32). In another study, Kalantari et al. (35) applied the PCR method to map E1 and E2 reading frames in HPV+ CC samples. They demonstrated that the 5’ end disruption was the most common pattern among different disruption/deletion patterns in the E1-E2 area of the HPV 16 genome in cervical carcinoma patients. The survival rate was poorer in patients diagnosed with carcinomas showing disruptions in E1/E2 than those without such changes. Concerning prognosis, E1 disruptions were considered critical (35). Contrary to the aforementioned studies in which combinations of therapeutic approaches were adopted, Lindel et al.’s study (33) was the first to include patients with HPV 16-positive uterine cervix cancer who were supposed to only receive radiotherapy as a curative modality. The results of their study are consistent with other studies demonstrating that disease-free survival is better in patients with intact E2 than in patients with disrupted E2.
In addition to the E2 gene status, another significant prognostic factor regarding the post-radiotherapy survival rate is the proportion of episomal to integrated copies of HPV DNA (29). According to a study by Shin et al. (29), cervical tumors with more episomal components are associated with better post-radiotherapy survival rates compared to purely integrated HPV DNA cells. In this study, ISH and the PCR technique were applied to differentiate between HPV DNA integration patterns. A multivariate model was used to categorize patients according to their different risk profiles. The best prognosis was found in tumors mainly composed of episomal HPV DNA, whereas the worst prognosis was detected in tumors with undetectable episomal DNA.
In a recent study by Zamulaeva et al. (36), radioresistance of CC cells was found to be dependent on both integration of HPV DNA into the host cell genome and amount of this integration. This study demonstrated that the percentage of cancer stem cells (CSCs) increased after irradiation in tumors with fully integrated HPV DNA whereas the proportion of CSCs decreased in tumors with episomal or partly integrated forms. This increase in the CSC pool after irradiation results from the subsequent proliferation of radio-resistant cancer cells and the differentiation of non-stem cells to stem cells as it occurs in radiation-induced epithelial-mesenchymal transition. Viral oncoproteins whose expression increases following HPV DNA integration into the host cell genome are known to enhance this transition (37).
What is fascinating is that the HPV DNA integration site, apart from the integration rate, appears to have a prognostic significance. Although HPV DNA integration is thought to be a random event, several studies have pointed out some hotspots that may affect cancer evolution (38-40).
In a study by Yang et al. (41), single-cell sequencing was employed to analyze mutations before and after radiotherapy in cervical tumors. They concluded that genetic characteristics were different in tumor cells pre- and post-radiotherapy. They concluded that genetic characteristics were different in tumor cells pre- and post-radiotherapy. Based on their results, tumor cells with HPV integration in POU5F1B, a well-known cancer susceptibility locus (42, 43), and a hotspot for HPV integration (44) are more likely to survive the following radiotherapy. This is assumed to be one of the possible underlying genetic mechanisms related to radiotherapy resistance.
4. Conclusions
Currently, radiotherapy alone or in combination with chemotherapy is the primary treatment for CC. However, the clinical outcome of treated patients is not the same as an endpoint survival rate after radiotherapy. Thus, it is necessary to identify potential biomarkers useful in personalizing therapy and optimizing radiotherapy outcomes. Several available reports have suggested that HPV integration patterns are an important and potentially useful clinical biomarker to guide high precision radiation therapy.