1. Background
Meat and its derivative products are considered as one of the most important food groups in a healthy diet, providing many essential valuable nutrients, including essential amino acids, beneficial fatty acids, and micronutrients such as iron, selenium, magnesium, zinc, and folic acid (1, 2). Given the nutritional significance of meat, a variety of meat products, either processed or unprocessed, from different domestic species of livestock and poultry such as cattle, sheep, camel, pig, chicken, ostrich, turkey, etc. are supplied in markets to provide customers' demands (3, 4). On the other hand, this need for meat products has tempted a few food manufacturers to engage in deceitful activities in the production process of meat products for obtaining more financial profits by using meat species different from what is labeled on the product packages (5). Therefore, this issue has recently raised consumers' concerns about the safety of meat products they eat and has called attention to the necessity of the development of meat authentication methods.
To identify meat authenticity, different types of excellent techniques such as near-infrared (NIR) technologies, chromatography, enzyme-linked immunosorbent assay (ELISA), and polymerase chain reaction (PCR) have been developed (6-8). In recent years, much attention has been paid to PCR-based methods, particularly real-time PCR, as sensitive, feasible, and reliable techniques for detecting the species origin of meats used in meat products (9-11). Recently, to increase the specificity and sensitivity of real-time PCR for simultaneously identifying two or more meat species in the minced meat samples or processed meat products, real-time PCR-high resolution melting (real-time PCR-HRM) was developed (11, 12). Relying on HRM analysis, the different PCR products specific to each meat species of interest would be distinguishable based on their differences in the sequence and GC content (13).
2. Objectives
Given the significance of meat authenticity and the fact that real-time PCR-HRM is known as a reliable method in this issue, we aimed to design our specific real-time PCR-HRM analysis and examine its validity to identify species-specific meat samples in both raw and cooked minced meat samples.
3. Methods
3.1. Sample Preparation
Meat samples from different species such as cattle, sheep, chicken, turkey, and wild pig were purchased from one of the reliable hypermarkets in Tehran, Iran. Meat preparation was performed according to the National Standards Organization of Iran No. 4622. Firstly, minced meat products with 10% and 20% purity of each selected species were prepared along with other food additives like onion, salt, spices, and toasted flour by mixing in a blender. Then, four different meat mixtures composed of four species-specific types of meat with 10% and 20% purity were prepared and divided into two groups of raw and cooked meat samples. The raw materials were stored at -20°C until required. In the cooked group, the mixed meat samples were cooked with fresh oil, and all hygiene considerations were addressed.
3.2. Sample Designing
Based on the meat source of each sample, the purity percentage of meat in the minced samples, and the raw or cooked forms of meat products, the prepared samples were classified into different groups (Table 1). The five types of the minced samples containing one meat type were named as A1 to A5, standing for 10% raw meat of cattle, sheep, chicken, turkey, and wild pig, respectively, A1+ to A5+ standing for 10% cooked ones, B1 to B5 standing for 20% raw meat of cattle, sheep, chicken, turkey and wild pig, respectively, and B1+ to B5+ standing for 20% cooked ones (not shown). Moreover, the mixed groups containing four different meat sources were categorized as follows: F1 and F2 stand for 10% and 20% of meat mixtures containing mutton, chicken, turkey, and beef, respectively, and F1+ and F2+ indicate the cooked form of F1 and F2 groups, respectively. Finally, H1 and H2 stand for 10% and 20% of meat mixtures containing wild pig meat, chicken, turkey, and beef, respectively, and H1+ and H2+ are defined as the cooked forms of H1 and H2 groups, respectively.
| The Mixed Samples | Cooking Status | Purity Percentage of Each Meat | Species |
|---|---|---|---|
| F1 | Raw | 10% | Mutton + Chicken + Turkey + Beef |
| F1+ | Cooked | 10% | Mutton + Chicken + Turkey + Beef |
| F2 | Raw | 20% | Mutton + Chicken + Turkey + Beef |
| F2+ | Cooked | 20% | Mutton + Chicken + Turkey + Beef |
| H1 | Raw | 10% | Wild pig meat + Chicken + Turkey + Beef |
| H1+ | Cooked | 10% | Wild pig meat + Chicken + Turkey + Beef |
| H2 | Raw | 20% | Wild pig meat + Chicken + Turkey + Beef |
| H2+ | Cooked | 20% | Wild pig meat + Chicken + Turkey + Beef |
3.3. DNA Extraction
The raw and cooked meat samples were subjected to DNA extraction by using a blood/tissue DNA extraction kit (DynaBio TM, Takapozist, Iran) according to the manufacturer's instructions. Briefly, the meat samples were chopped and slashed by scalpel and transferred to 1.5-mL microtubes. For lysing the cells and releasing the genomic DNA, 300 µL of lysis buffer (TL) and 20 µL of proteinase K were added to samples and rigorously vortexed for 20 sec. The resulting lysates were centrifuged at 12000 rpm for one minute to separate genomic DNA from cell debris. In the next step, 200 µL of supernatant was transferred to a new microtube, and 200 µL of binding buffer and 200 µL of absolute ethanol were added, and they were mixed thoroughly. The whole mixture was transferred into a binding column loaded on a collection tube (all provided by kit) and centrifuged at 5000 rpm for one minute. DNA molecules trapped on the binding column were washed twice with washing buffers 1 and 2, respectively, and then centrifugation at 8000 rpm was performed for one minute. Finally, the purified genomic DNA was eluted in 60 µL of elution buffer and collected in a new microtube. All the DNA samples were stored at -20°C until used.
3.4. Real-time PCR Assay
To recognize each species-specific meat in the mixed samples at the molecular level, a region of cytochrome b gene located in the mitochondrial DNA was chosen. The cytochrome b sequences of all five species were aligned, and the species-specific regions were addressed for designing the species-specific primers. The specific primers for each meat type (i.e., wild pig meat, chicken, beef, mutton, and Turkey) were designed according to the NCBI database (Table 2). Real-time PCR was performed using Qiagen’s real-time PCR machine (Rotor-Gen Q). The PCR reaction mixtures were composed of 2 µL of DNA sample, 2.5 µL of a qPCR master mix containing Cyto 9 fluorescent dye (Life Technologies Corp., Carlsbad, CA), 1 µL of MgCl2, 2 µL of dNTP, 0.5 µL of each primer (10 mM), 0.2 µL of Taq polymerase enzyme, and 16.5 µl of ddH2O. The PCR thermal process was programmed as follows: an initial denaturation step at 94°C for 5 min, followed by an amplification step including 35 cycles of 94°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec.
| Samples & Primers | Sequences (5´- 3´) | Tm (°C) | Amplicon Size (bp) |
|---|---|---|---|
| Universal forward primer | CCTCCCAGCTCCATCAAACATCTCATCTTGATGAAA | ||
| Reverse primer | |||
| Beef | CTAGAAAAGTGTAAGACCCGTAATATAAG | 84.7 | 274 |
| Mutton | CTATGAATGCTGTGGCTATTGTCGCA | 82.8 | 331 |
| Chicken | AAGATACAGATGAAGAAGAATGAGGCG | 87.8 | 227 |
| Turkey | AAGATACAGATGAAGAAGAATGAGGCG | 85.7 | 227 |
| Wild pig meat | GCTGATAGTAGATTTGTGATGACCGTA | 83.8 | 396 |
3.5. High-resolution Melting Analysis
At the end of real-time PCR, HRM analysis was performed by gradually raising the temperature from 72°C to 95°C at a rate of 0.2°C/s. The resulting melting profiles related to each meat type were obtained using Rotor-Gene 1.7 software. Normalization regions of 81-89.5 were considered for analyses, and the identical graphs were classified into the same group with a 95% confidence percentage.
3.6. Statistical Analysis
All the experiments were performed in duplicate. The resulting data were analyzed by Excel (version 2017) and presented as mean.
4. Results
4.1. Identification of Species-Specific Cytochrome B Fragments by Real-time PCR-HRM Analysis
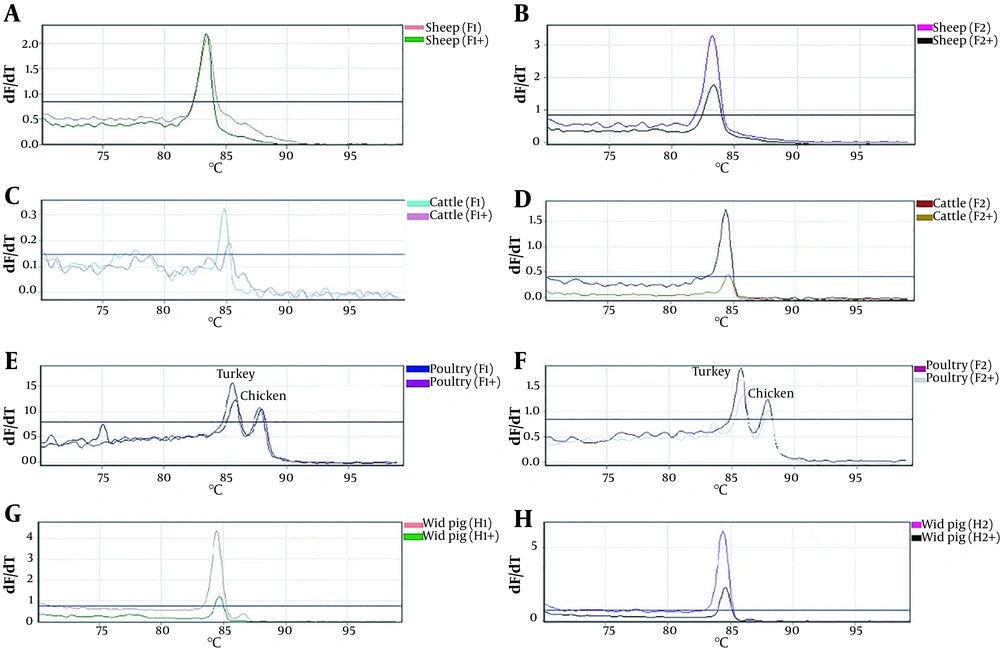
The real-time PCR amplification and HRM analysis were performed to detect cytochrome b DNA fragments specific to each species of interest (i.e., cattle, sheep, chicken, turkey, and wild pig). As shown in Figure 1, the species-specific DNA fragments were successfully amplified by real-time PCR, and their specific melting curves validated the specificity of PCR product of each species, presenting a single melting curve with a specific melting temperature. In the case of chicken and turkey, despite using the same couple of primers, two distinct melting curves (85.7°C for turkey and 87.8°C for chicken) were obtained (Figure 1E and F ). The melting temperatures of species-specific PCR products were presented as follows: sheep: 82.8°C; cattle: 84.7°C; chicken: 87.8°C; turkey: 85.7°C, and wild pig: 83.8°C.
Identification of species-specific cytochrome b fragments by real-time PCR-HRM. The specific melting curve related to each meat species was obtained by the PCR amplification of cytochrome b fragment using species-specific primers. The melting temperature of the species-specific amplified fragments was presented as follows: sheep: 82.8°C; cattle: 84.7°C; chicken: 87.8°C; turkey: 85.7°C, and wild pig: 83.8°C.
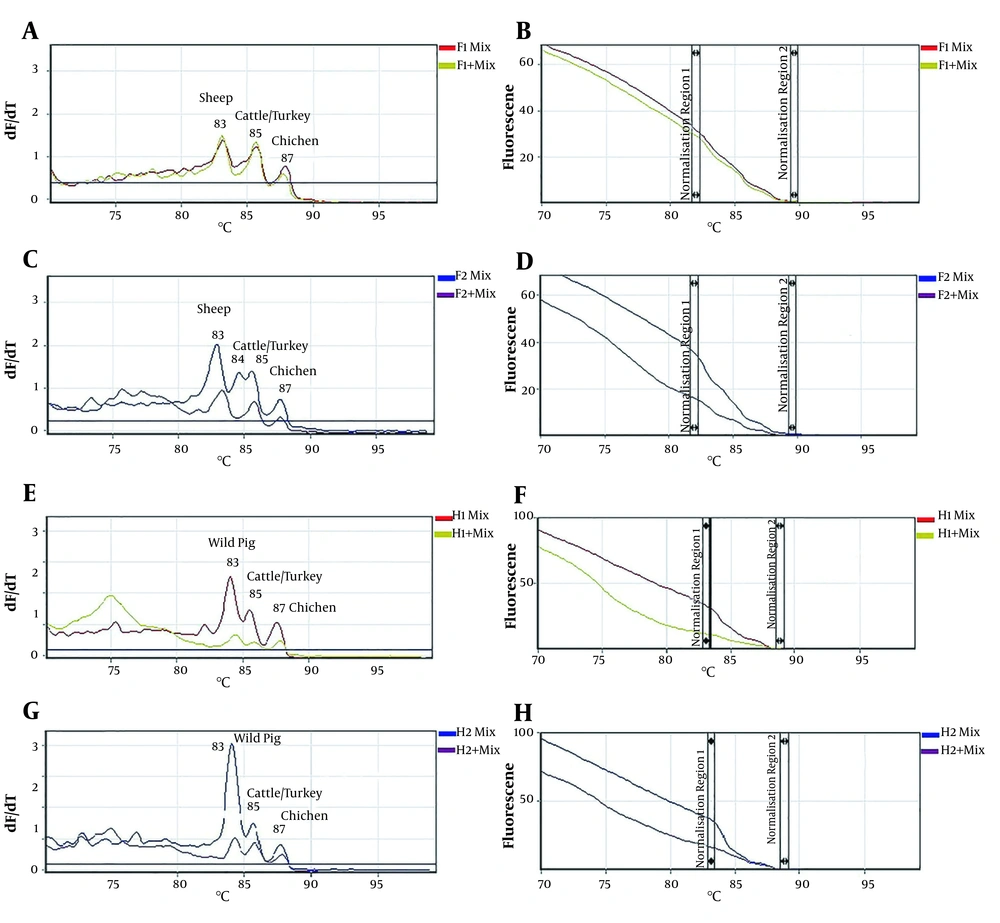
4.2. Detection of Different Species in the Mixed Meat Samples Using HRM Analysis
The results obtained from real-time PCR-HRM analysis of the mixed meat samples indicated that species-specific DNA existing in different meat mixtures was successfully amplified and distinguished via its specific melting temperature. All the meat mixtures (including raw and cooked, 10% and 20% purity) showed distinct melting curves with specific Tm for each species (Figure 2 A, C, E, and G). As presented, the resulting melting curves showed three peaks at 83°C, 85°C, and 87°C for sheep, cattle/turkey, and chicken samples of F1 and F1+ groups, respectively. Three peaks of melting curves at 83°C, 85°C, and 87°C for wild pig, cattle/turkey, and chicken samples were also observed in H1, H1+, H2, and H2+ groups, respectively. Due to the Tm similarity of the amplified DNA fragments related to cattle and turkey samples, their melting curves were not distinguishable in multiplex PCR of the mixed samples; however, they were detected as almost separated peaks in F2 and F2+ groups. Furthermore, the resulting HRM curves of the four used meat mixtures (F1, F2, H1, and H2) also demonstrated several temperature shifts related to each species-specific meat DNA (Figure 2B , D, F, and H).
5. Discussion
Increasing fraud in meat products has recently become an important concern of consumers all over the world (5). Therefore, meat authentication has been addressed as a critical issue that is directly associated with community health. On the other hand, identifying meat species in different meat products is particularly considered as an important issue in Islamic countries where people only consume Halal meats (14). In the last decades, PCR-based methods have been widely used to determine the meat authenticity of various food products (raw, precooked, and cooked) provided by different animal species (9, 15).
In this study, the real-time PCR technique with HRM analysis was used for meat authentication of the raw and cooked meat samples composed of four different meat species. The HRM analysis as a highly sensitive method allows discriminating several meat samples in a mixture. By real-time PCR-HRM analysis, we could distinguish the meat species in the mixed meat samples. Our results confirmed the high sensitivity of real-time PCR-HRM assay and validated the use of cytochrome b gene as a suitable mitochondrial gene for detecting different species-specific DNAs in the meat mixtures.
Several previous studies have also utilized species-specific PCR assays through amplifying species-specific mitochondrial DNA regions to identify poultry residuals in beef sausage, buffalo meat in hamburgers, or squid species in mixed samples (10, 16, 17). Consistent with previous studies, our data also showed that raw or cooked meats had no significant impact on PCR efficiency (18). Our previous study (Unpublished work) showed that qPCR-HRM analyses are adequately sensitive to detect one and two different meat species in the minced meat samples. Moreover, we showed in this study that the qPCR-HRM method also exhibited high efficiency for the detection of four different meat species in meat mixtures. We successfully detected each meat species with its specific Tm in the mixed samples, while the real-time PCR-HRM was performed using each pair of species-specific primer separately.
Jonker et al. (2008) also developed a real-time PCR method to identify meat species in mixture samples containing beef, pork, horse, mutton, chicken, and turkey. By adjusting the real-time PCR program, they showed that it was possible to detect meat species at a level of 0.05% in test mixtures. However, there might be some cross-reactivities between the species of interest, they demonstrated that amounts as high as 250 ng DNA of the species showed no false signals (15). In multiplex HRM analysis, we also observed that the resulting melt curves for all the groups (except for F2 and F2+) illustrated three peaks rather than four peaks indicative of four meat species used in meat mixtures. This observation seems to be due to the Tm proximity of the amplified DNA fragments related to cattle and turkey samples.
Hence, to achieve higher specificity, it is necessary to refine primer designing to amplify fragments with different Tms, so that they will be distinguishable in the mixed meat samples. Furthermore, as in various other studies, the TaqMan probe-based real-time PCR method could be used as a more sensitive technique to discriminate each meat species DNA by TaqMan probes specifically designed for each species (19, 20). Consequently, as previous studies reported, the real-time PCR showed a better performance compared to other procedures.
In addition to meat authentication, many studies have reported that the real-time PCR-HRM method would be a reliable method for identifying halal food authentication, which is an important issue in Islamic countries (21). Consequently, the data presented here demonstrated that the real-time PCR-HRM assay would be a promising tool for the species identification of meat products. Considering the increasing methods for adulteration detection, development of high technologies for more sensitively detecting any meat contamination in processed foods is required.
5.1. Conclusions
In sum, the current study indicates that the real-time PCR-HRM method could be considered as a reliable technique for meat authenticity detection of the processed products and distinguishing halal and haram meat samples; however, some refinements are required for improving the specificity of these methods.