1. Background
Cancer is one of the major public health issues and the leading cause of death in humans (1). Numerous studies have shown that cancer often occurs due to alterations in the balance of oncogenes and tumors suppressor genes. MicroRNAs (miRNAs) are essential nucleotide structures in a number of pathways associated with physiological and pathological processes, including proliferation, cell cycle, migration, and cell apoptosis (2, 3). Recent studies have identified miRNAs as a new regulator of oncogenes and tumor suppressor genes. It is believed that miRNAs can be considered a new method for cancer diagnosis and treatment (4). Also, miRNAs act as oncogenes or tumor suppressors through various mechanisms such as modifying proliferation signaling, growth suppressors escape, resisting cell death, activating invasion and metastasis, and inducing angiogenesis (5).
MiRNA mimics and inhibitors are currently in clinical practice and have shown promise as new therapeutic agents (4). MicroRNAs are secreted into exosomes or microvesicles. These secreted structures are highly stable and can be absorbed by surrounding tissue cells or migrate to distant sites through circulation, resulting in the reduction of the expression of their target proteins by reducing target mRNA levels (2, 6, 7). Glioblastoma multiforme (GBM), also known as grade IV astrocytoma, is a fast-growing invasive brain tumor that attacks nearby brain tissue. Glioblastoma is the most common neoplasm of the central nervous system and accounts for 10 - 15% of all cases of central nervous system tumors. Combination therapy involving surgery, along with chemotherapy or radiotherapy, is currently being used. However, due to the rapid growth of GBM, high invasiveness, and lack of drug resistance, patients usually have a poor prognosis, and the average survival rate is 14.6 months (2, 6).
MiRNAs play a vital role in several biological processes in glioblastoma, including cell growth, migration, invasion, angiogenesis, and stem cell-like behavior (8, 9). Regulatory miRNAs have been recognized as potential therapeutic molecules. For example, miR-34a, miR-124, miR-128, miR-137, miR-181, and miR-7 families are only a few of the miRNAs with low expression in GBM. Recent studies have pointed to the significant reduction in expression of the miR-7, miR-34a, miR-124, miR-128, miR-137, miR-181, and miR-627 families in several tumors, including glioblastoma. In contrast, the expression of the miR-17-92 cluster, which includes miR-18a, miR-19a, miR-19b, miR-20a, and miR-92a, is increased in GBM and results in the silencing of various target genes involved in processes such as cell cycle cessation, angiogenesis, and apoptosis, and has important functions in gliomagenesis. This category includes miR-10b, miR-15b, miR-26a, miR-17-3p, miR-17-5p, miR-93, miR-148, miR-182, and miR-21. These oncomiR target downstream genes and are involved in proliferation, cell survival, invasion, and therapeutic resistance. Other miRNAs with increased expression in glioblastoma include miR-10b, miR-15b, miR-26a, miR-93, miR-148, miR-182, and miR-221/222, which play key roles in the development of glioblastoma (7).
2. Objectives
3. Methods
3.1. Study Population
A total of 50 patients with suspected GBM referred to Imam Khomeini and Shohada of Tehran Hospital were evaluated. Of them, 29 men and 21 women were included in the study. Also, a very small amount of healthy tissue around the tumor was used as a normal sample. In this study, pathological methods were used to diagnose healthy and patient samples. The age range of the patients was between 28 - 76 years (mean 52 and standard deviation 0.3694). Patients' demographic data were collected through face-to-face interviews and medical records. All subjects who participated in the study were provided with written informed consent and assured that no financial or human loss would be incurred during the study. None of the patients had a family history of GBM cancer. The process was approved by the Ethics Committee of Islamic Azad University North Tehran Branch (Ethics code: IR.IAU.TNB.REC.1400.051).
3.2. Sample Preparation
Total RNA was extracted using mirVana ™ PARIS (Ambion) kit to evaluate the expression of miR-4772-3p and miR-3173-3p genes in patients and healthy individuals. To this aim, 2-mercaptoethanol was added to the samples at room temperature (RT) at a 1: 1 ratio. Then acid-phenol: chloroform solution was added to the resulting mixture at a 1: 1 ratio and vortexed for one minute. Then the mixture was centrifuged for five minutes at 10,000 g at RT. The aqueous phase was transferred to a new 1.5 mL microtube and 400 µL absolute ethanol was added and mixed well. The resulting solution was transferred into a silica filter column and centrifuged for 30 seconds. In the next step, 700 µL washing solution-1 was added to the filter and centrifuged for 15 seconds. The previous step was repeated by adding 500 µL absolute ethanol and centrifugation for one minute without adding solution. Finally, the filter was placed on a new 1.5 mL microtube and 100 µL nuclease-free water at 55°C was added to the center of the filter and incubated for five minutes at RT. The filter was then centrifuged for 30 seconds and stored at -80°C for following analyses. The purity of the extracted RNA was evaluated using the optical density (OD) of the RNA solution at 260 and 280 nm by a NanoDrop device (Q6000 B) and the optimum 260 nm/280 nm ratio of 1.2/1.8 was obtained. Electrophoresis on a 1.5% agarose gel was also used to ensure the quality and purity of the extracted RNA.
3.3. cDNA Synthesis
cDNA synthesis was done using a cDNA synthesis kit (Aess 1211-2). Microtubes containing RNA and cDNA synthesis enzymes were incubated at 37°C for 15 minutes and then at 85°C for 15 seconds in the thermocycler. Synthesized cDNA was stored at -20°C until Real-time PCR.
3.4. Primer Design and Synthesis
The primers for miR-3173-3p, miR-4772-3p, and GAPDH were designed using Gene runner software and synthesized by Pishgam Biotech Company. The sequences of the primers are as follows: miR-3173-3p F: GGGAAAGGAGGAAATAGGC, miR-3173-3p R: GTGCAGGGTCCGAGGT and miR-4772-3p F: GTTCCTGCAACTTTGCCTG, miR-4772-3p R: GTGCAGGGTCCGAGGT.
3.5. Real-time PCR
The expression of the genes of interest was evaluated by SYBR green method using Step One, Applied Biosystem thermocycler, Singapore. To ensure the specific amplification of the fragments, the melt curve of the miR-3173 and miR-4772-3p genes were also analyzed. The procedure was done according to the kit protocol (Amplicon Denmark), and the reactions were subjected to the following thermal program for 40 cycles: 30 seconds at 95°C (Activation), 5 seconds at 95°C (Deactivation), and 30 seconds at 58°C (Primer Annealing and Extension). The relative expression of the target gene was analyzed using the -∆∆ct formulae. The GAPDH gene was used as the reference (housekeeping) gene for relative quantification (RQ) of the target genes. The experiments were done in duplicate.
3.6. Statistical Analysis
Descriptive data were analyzed using GraphPad Prism version 8 and parametric data were analyzed using Oneway-ANOVA and T-test. For data that do not show Gaussian distribution, Mann-Whitney and non-parametric Kolmogorov-Smirnov and Shapiro-Wilk tests were used. P-value ≤ 0.05 was considered significant.
4. Results
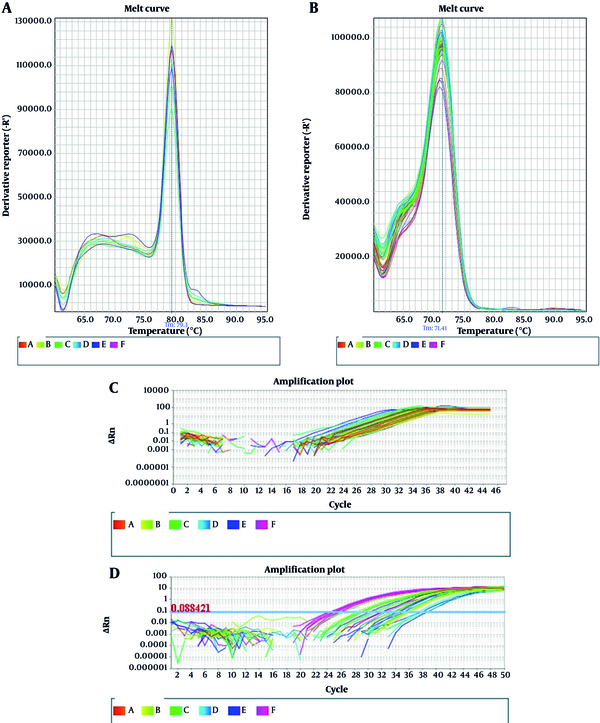
Real-time PCR results are displayed in Figure 1. Melt curve analysis was used to examine the results of amplification of specific fragments of miR-3173-3p and miR-4772-3p genes and possible formation of primer dimers. As shown in Figure 1A-B, both genes are specifically propagated. Figure 1C-D depicts the typical amplification plot obtained from SYBR green real-time PCR with glioblastoma cancer tissue. The horizontal line corresponds to the threshold, which is defined at the level of the highest amplification rate during the initial exponential phase.
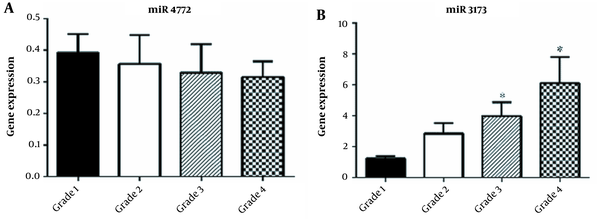
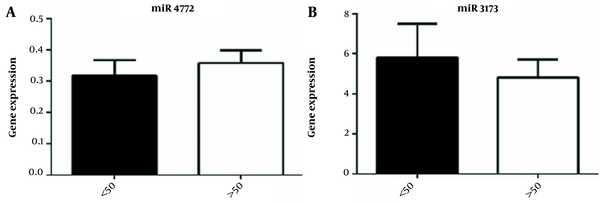
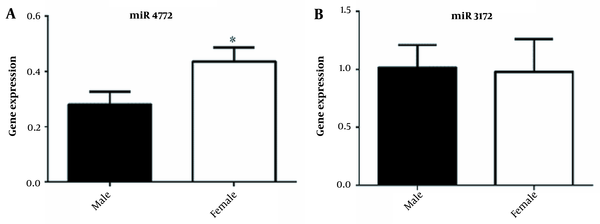
Analysis of the expression of miR-3173-3p and miR-4772-3p genes in the course of the disease showed that the expression of miR-3173-3p increased with progression of the disease and was statistically significant in grades III and IV of the disease (Figure 2A-B). No significant correlation was observed between the age of participants and the expression of miR-3173-3p (P-value = 0.57) and miR-4772-3p (P-value = 0.56) (Figure 3A-B, Table 1). Also, as shown in Figure 4A-B, there was no significant correlation between sex and the expression of miR-3173-3p gene (P-value = 0.91), while miR-4772-3p expression was significantly higher in women with GBM compared to men (P-value = 0.03).
| Variables | Glioblastoma | Healthy | Total | Mean RQ of miR3173-3p | Mean RQ of miR4772-3p |
|---|---|---|---|---|---|
| Number of patients | 46 | 50 | 100 | ||
| Age | |||||
| < 50 | 14 | 20 | 36 | 5.830 | 0.3173 |
| > 50 | 32 | 30 | 64 | 4.815 | 0.3579 |
| Sex | |||||
| Male | 33 | 30 | 59 | 1.015 | 0.2809 |
| Female | 19 | 20 | 41 | 0.9790 | 0.4351 |
| Grade | |||||
| I | 14 | - | 14 | 1.235 | 0.3929 |
| II | 11 | - | 11 | 2.842 | 0.3571 |
| III | 9 | - | 9 | 3.991 | 0.3295 |
| IV | 11 | - | 11 | 6.107 | 0.3161 |
The Correlation Between Age, Sex, and Disease Parameters with the Expression of miR-3173-3p and miR-4772-3p
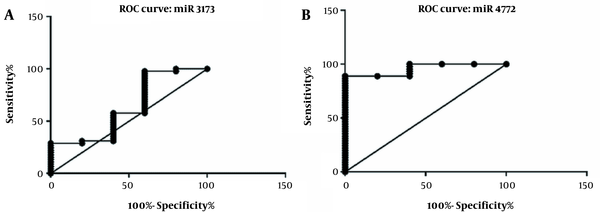
Biomarker evaluation of miR-3173-3p and miR-4772-3p was assessed using ROC curve, as shown in Figure 5A-B. The area under the curve shows the sensitivity and specificity in differentiating GBM patients from healthy individuals. The area under the curve of miR-3173-3p was 0.6311, and its significance level was 0.34 with CI = 0.3475 to 0.9147. The area under the curve for miR-4772-3p was 0.9556, and its significance level was 0.0009 with CI = 0.8921 to 1.019. Although the ROC curve evaluation of miR-3173-3p showed that the expression of this gene failed to differentiate between healthy and diseased groups, the ROC curve for miR-4772-3p expression could effectively differentiate between the patients and healthy controls (Figure 5A-B). Determination of cut-off value showed that miR-4772-3p, with cut-off of 0.6939, differentiated GBM patients from healthy individuals with 100.0 specificity and 88.89 sensitivity.
5. Discussion
So far, a large number of miRNAs have been identified as key regulators of carcinogenesis (10, 11). In the present study, Real-time PCR was used to compare the expression profiles of two miRNAs, including miR-3173-3p and miR-4772-3p, in GBM patients and healthy individuals, and an integrated molecular network for identifying potential target miRNA interactions in glioblastoma carcinogenesis was proposed.
In a study, clinical signs of pathology and expression of six microRNAs, including miR-3173-3p, were evaluated in patients with tumor deposits. TDs (Tumor deposits)-related miRNAs were downregulated (12). Also, its downregulation exacerbates cell invasion in B-cell acute lymphoblastic leukemia (B-ALL) (13). In contrast, the expression of miR-3173-3p in grade IV GBM increased compared to grade III. Also, in some patients with ovarian cancer, low expression of NF90 and high miR-3173-3p expression reduce their survival. Also, overexpression of miR-3173-3p increases metastasis with the help of NF90 and DICER molecules (14).
Small nucleolar RNA host gene 3 (SNHG3) has been reported as oncogenic long noncoding RNA in many cancers. SNHG3 was significantly upregulated in CCA (cholangiocarcinoma) cells compared with normal human intrahepatic biliary epithelial cells. Knockdown of SNHG3 inhibited the proliferation and migration of CCA cells. Mechanistically, SNHG3-sponged miR-3173-5p, thus releasing the repression of ERG by miR-3173-5p. Rescue experiments showed that miR-3173-5p/ERG axis mediated the oncogenic effect of SNHG3 (15).
The acting of miR-3173 in B-ALL has been previously investigated; however, its real function in the progression or suppression of the disease has not yet been revealed. Further studies showed a significant decrease in the expression of miR-3173 in B-ALL. The results showed that miR-3173 inhibited cell proliferation, migration, and invasion of SUP-B15 and CCRF-SB cells, whereas miR-3173 suppressed these cellular functions. Since proliferation, migration, and invasion are key elements of cancer progression, the research showed that miR-3173 played an important role in the pathogenesis of B-ALL and could be considered a therapeutic approach for this disease. MiR-3173 also inhibits PTK2. This miR-3173/PTK2 axis provides new insights into the underlying mechanisms of B-ALL (13). Another study on ovarian cancer patients showed that low expression of NF90 and high expression of miR-3173-3p could be used as independent prognostic markers of poor survival in this malignancy (14).
Regarding the role of miR-4772-3p in cancer pathogenesis, studies have reported a significantly decreased expression of miR-4772-3p in patients with recurrent colon cancer, and it can be considered as one of the potential markers of the disease. The studies have also suggested a different expression pattern of miR-4772-3p in breast cancer compared to normal tissues (16) and in patients with any levels of Alzheimer's disease (17). Although the function of miR-4772-3p remains unclear, several genes have been identified as its targets, including RTN4 and RABA9A. RTN4 protein, also known as NOGO, has different functions in epithelial–mesenchymal transition, cell adhesion, and migration in lung cancer (18), hepatocellular carcinoma (19), and cervical cancer (20). The major molecule of ras oncogene superfamily is RAB9A gene (21). Reduction in serum level miR-4772-3p is an important biomarker for colon cancer. (22). In this regard, the study of miR-4772-3p expression shows that the disease stage has no significant effect on the expression of this microRNA.
Analysis of the expression of miRNAs such as miR-4772-3p in the pathogenesis of sepsis suggests that they can be used as biomarkers for the diagnosis and prognosis of sepsis (23). Bioinformatics analysis in pancreatic cancer showed that miR-4772-3p with a downregulating mechanism had a great potential to be used as a prognostic factor and therapeutic target for this tumor (24). Also, in this study, the expression level of miR-4772-3pwas performed in comparison with normal tissues with Real-time PCR method on glioblastoma cancer tissues. We found that age and stage of the disease did not have a significant effect on the expression of this gene.
5.1. Conclusions
According to the results of the present study, miR-3173-3p was produced by glioblastoma tumor tissues. Also, the serum levels of miR-3173-3p in grades III and IV patients were significantly higher than healthy individuals and might help differentiate between the two groups. The expression of miR-3173-3p in serum was also increased with the progression of the tumor grade. Therefore, serum miR-3173-3p levels could be considered a non-invasive method for early diagnosis of patients with GBM.