1. Background
Systemic lupus erythematosus (SLE) is a complex autoimmune disease with unknown etiology, characterized by immunoglobulin autoantibody production against nuclear, cytoplasmic and cell-surface auto antigens, resulting in inflammation and tissue damage (1). There are a number of environmental triggers and genetic factors causing this disease. Some environmental and hormonal factors may be conducive to its pathogenesis, such as the infectious agents, diet, toxins/drugs, and ultraviolet (UV) light (2, 3). Genetics is also a factor determining a person’s propensity to develop SLE; so, analyzing the genes and molecular interactions that influence the disease can promote our understanding of the pathogenesis and genetic contributions to autoimmunity in SLE (4). Study of genetic associations and analysis of single nucleotide polymorphisms (SNPs) have contributed to the identification of several loci associated with the disease susceptibility (5, 6). Major histocompatibility complex (MHC) gene family is known as an outstanding genetic factor of autoimmune diseases in humans. One of the genes reported to be associated with autoimmune diseases such as SLE is CTLA4. This gene consists of four exons and the size spans to 6,175 bases, with a molecule highly similar to the CD28 molecule (7). The genes encoding CTLA4 and CD28 cell surface receptors expressed by T cells are in region 2q33, identified as a susceptibility region for SLE in genome-wide scans (8, 9). Improper T-cell-dependent expansion of auto-reactive B and T cells has been considered to play an important role in the SLE pathogenesis. For activation of T-cells, MHC class II molecules on the surface of antigen-presenting cells are recognized by CD4 T-cells, but recognition is not sufficient and co-stimulation by other receptor–ligand complexes is required. CD28 and CTLA4 receptors and their ligands B7-1 (CD80) and B7-2 (CD86) are the main co-stimulatory molecules involved in this system. CTLA-4 can regulate T-cell polarization to T-helper (Th) 1 or Th2 by controlling the overall strength of T-cell activation signal (10). Some polymorphisms have been reported in the CTLA4 gene, possibly associated with SLE susceptibility, of which an A/G transition at position-1661 within the promoter region is one of the most important variations (11-13). This polymorphism may apply alterations in the potential response element for myocyte enhancer factor 2 (MEF2) (14). Hence, allelic variations of this site may cause susceptibility to SLE, resulting from unbalanced or inefficient immune responses. Although CTLA-4 polymorphism has been detected to be associated with a number of autoimmune diseases including SLE, Graves’ disease, type 1 diabetes, and multiple sclerosis, yet the results of associations are different between populations (11, 12, 15).
2. Objectives
Recent studies have shown that CTLA-4 polymorphism plays an important role in SLE in some populations, which has not been confirmed in Iran. We used a case-control study to investigate the role of CTLA-4 polymorphism at position-1661 in SLE susceptibility in our Iranian population with SLE.
3. Patients and Methods
3.1. Patients with SLE
We studied 31 unrelated patients with SLE (27 females and four males) with mean age of 11.35 years (ranging from 6 to 16) and 50 unrelated healthy subjects (24 females and 32 males) as controls. Patients were recruited from Mofid Hospital, Tehran, IR Iran. They were recognized according to the American College of Rheumatology criteria (16). The Healthy subjects did not have clinical evidences or history of autoimmune and genetic diseases. The study was confirmed by the local ethics committee and all subjects voluntarily participated in the study.
3.2. Samples and Genotyping
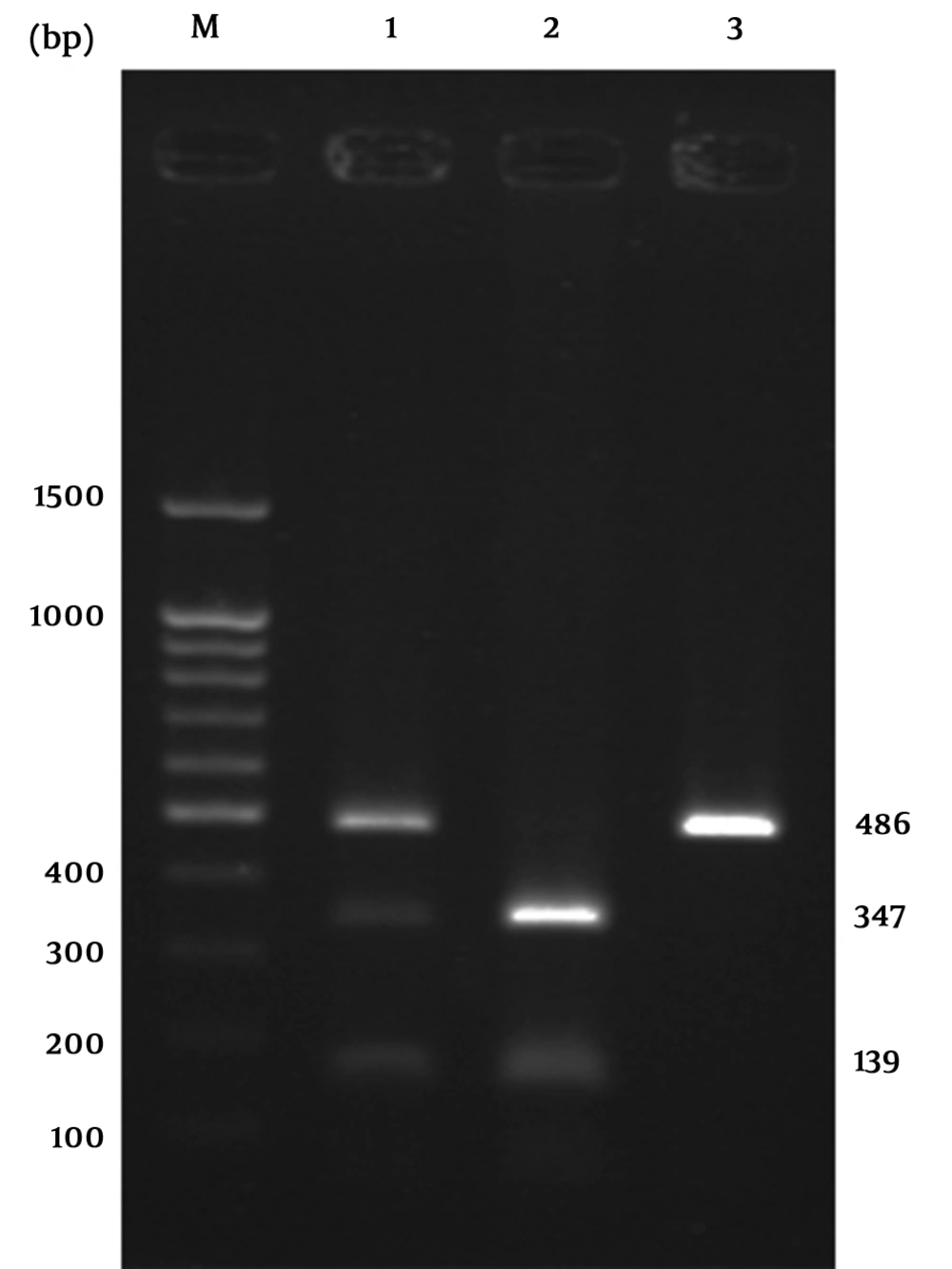
We extracted the genomic DNA from the anti-coagulated whole blood of each patient using the Saremi et al. protocol (17). DNA extraction kit was also used for some samples according to the manufacturer’s recommendations (Qiagen, Hilden, Germany). Polymerase chain reaction (PCR) was performed for each sample in a total volume of 25 µL using 2 - 6 µL of genomic DNA, 1 µL of primers (Forward primer: 5ʹ CTAAGAGCATCCGCTTGCACCT 3ʹ and reverse primer: 5ʹ TTGGTGTGATGCACAGAAGCCTTTT 3ʹ), 2.5 µL of 10x buffer (cinnagen, Iran), 0.5 µL of dNTP (Geneon), 0.5 µL of Taq polymerase (Kowsar, Iran) and 0.75 µL of MgCl2 (cinnagen, Iran). The PCR-RFLP method was used for analyzing the polymorphisms at position-1661. The study followed optimal PCR conditions, an initial 94°C denaturations for 5 minutes followed by 40 cycles of denaturation at 94°C for 30 seconds, annealing at 58°C for 30 seconds, elongation at 72°C for 1 minute and one final extension at 72°C for 10 minutes. Afterwards, the PCR products (486 bp) were run on 1% agarose gel. Low-range DNA ladder (Takara, Tokyo, Japan) was used to confirm the results. PCR products were digested with 0.2 µL MseI enzyme (Vivantis, Malaysia) at 60°C for 1 hour and then run on 2% agarose gel. The 1661 A/G polymorphism was identified by detecting a 486 bp fragment (G allele) or two fragments of 347 and 139 bp (A allele) (Figure 1).
3.3. Statistical Analysis
To compare the genotypes and allele frequencies between patients with SLE and controls, χ2 and Fisher's exact tests were used. Each allele frequency in patients with SLE was compared with the same allele in controls; P < 0.05 was considered significant. The odds ratio (OR) and 95% confidence intervals (CI) were calculated using 2 × 2 contingency tables to measure the association of particular alleles and genotypes. SPSS statistical software version 18 was used for all statistical analyses.
4. Results
All samples from 31 patients with SLE and 56 healthy controls were genotyped for the 1661 A/G polymorphism in the CTLA-4 gene. As it is indicated in Table 1, there were no differences in the allele or genotype frequencies of this polymorphism between patients with SLE and controls. Comparison of allele and genotype frequencies in case and control groups showed a higher frequency of A allele as well as AA genotypes in both case and control groups.
| -1661 Promoter | Patients, n = 31, No. (%) | Controls, n = 50, No. (%) | χ2 | OR (95% CI) | P Value |
|---|---|---|---|---|---|
| Genotype frequency | |||||
| AA | 20 (64.5%) | 30 (60%) | 0.533 | 1.238 | 0.465 |
| AG | 10 (32.2%) | 20 (40%) | 1.389 | 0.706 | 0.239 |
| GG | 1 (3.3%) | 0 | 3.046 | 0 | 0.081 |
| Allele frequency | |||||
| A | 50 (80.6%) | 80 (80%) | 0.032 | 1.066 | 0.858 |
| G | 12 (19.3%) | 20 (20%) | |||
| Phenotype | |||||
| A | 30 (96.7%) | 50 (100%) | 0.144 | 1.109 | 0.704 |
| G | 11 (35.4%) | 20 (40%) |
Genotypic and Allelic Frequencies of -1661 CTLA-4 Polymorphisms in Iranian Patients with SLE and Healthy Controls a
5. Discussion
According to our results, there were different outcomes for various populations. Several studies have shown a significant association between SLE and CTLA-4 gene polymorphisms (12, 13, 18, 19). However, other studies demonstrate no association with this genetic variation (20, 21). Reasons for variation in genetic susceptibility are still ambiguous. Hence, investigations on the frequencies and distributions of CTLA-4 gene variations across populations are necessary for understanding the disease mechanisms and significance of CTLA-4 gene in SLE; CTLA-4 locus on chromosome 2q33 has been reported to show linkage with SLE (22, 23). This polymorphism has not been investigated in the Iranian population, but the association has been observed in multiple independent studies of different Asian populations (12, 13, 18, 24) and there is strong evidence showing that CTLA-4 polymorphism confers susceptibility to SLE due to its crucial functions in T-cell activation and regulation (11, 23, 24). In our study, allele frequencies for the -1661 site were not found to be significantly different between patients and controls. In another study on a Korean population with SLE -1661 polymorphism, positive correlation was not indicated (13). In addition, no associations were found between this polymorphism and SLE in African-Americans (21). However, Subgroup analyses of African-American youth (< 35 years) revealed effect modification by age for the -1661 G allele, showing a significant positive association with SLE (21).
CTLA-4 polymorphism at position -1661 was not significantly associated with patients with SLE in our study. Our results were in accordance with the majority of published studies supporting the important influence of CTLA-4 polymorphisms in susceptibility to SLE. Nevertheless, further functional studies are required for analyses of polymorphisms on the CTLA-4 gene, and larger population studies should be performed in different subgroups in order to fully examine the SLE associations with the CTLA-4 locus. Frequency of alleles and genotypes may differ between populations; therefore, association studies of this gene present a possible route for understanding the influence of genetic variations in this gene on the disease and help in developing new treatments.