1. Background
Colorectal cancer is the third most common cancer in the world and the forth cause of death globally. It usually starts as a benign tumor, often a polyp, which becomes cancerous within some years (1). Colorectal polyps are classified into 2 major groups: conventional adenomas and serrated polyps (2). Neoplastic polyps of the colorectal tract, such as tubular and villous adenomas, are considered precursor lesions leading to colorectal cancer (3). Sporadic polyp is a disease of the intestines afflicting people in the world and is associated with an increased risk of colorectal cancer (CRC) (4, 5). DNA methylation changes in colonic epithelial cells that normally occur with aging are accelerated in patients with polyp and adenomas colon cancer, because of higher cell turnover in the inflammation (6). In individuals over the age of 50 years, the prevalence of hyperplastic polyps is 20% - 40%. Hyperplastic polyps could act as a marker for future adenomas in the general population (7). Furthermore, the sequence of serrated polyp carcinoma is recently suggested as another pathway of colorectal-carcinogenesis to the previously known adenoma-carcinoma sequence (2). Due to difficulty in colonoscopy method, also the need to a lot of colonic biopsies and rather low accuracy to recognize early detection of polyp and adenoma associated carcinoma, there is a great inquiry to find reliable molecular markers to early detection of polyp-associated neoplastic lesions (8). Among these molecular markers, epigenetic changes, especially DNA methylation of cancer related genes, are very important and early event (3, 9). In spite of genetic alterations, epigenetic modifications including CGI (CpG Island) DNA methylation, also occur in colon polyps and colon cancer (10); CGI DNA methylation is an epigenetic mechanism that represses gene transcription in normal cellular processes, but becomes excessive and aberrant in many neoplasms (11). DNA methylation is a ‘‘second hit'' mechanism in CRC and characterizes the role of DNA methylation in the polyp phase of colorectal cancer (12). Frequent promoter methylation and CGI methylator phenotype (CIMP) are reported in serrated polyps; however, recent studies show high frequency of promotor methylation and CIMP in conventional adenomas (2, 11, 13-15). Powerful signaling pathways such as Wnt signaling are controlled by negative regulations such as soluble Fz-related proteins (SFRPs) and Wnt inhibitory factor (WIF1) that normally inhibit Wnt signaling pathway by binding to its extracellular ligands (13). Inactivation mechanisms lead to silencing SFRPs and WIF1, and such as promoter methylation can cause aberrant activation of Wnt signaling in cancer cells (13, 14). Moreover, MGMT deficiency is likely to be responsible for the emergence of MSI CRCs in different clinical contexts (15). Frequent methylation of MGMT is detected in colorectal tumor tissues; however, minimal MGMT methylation is found in tissues of healthy people (16). Methylation of MGMT promoter is reported with equal frequency in small adenomas, large adenomas, and carcinomas; indicating that these changes occur early in neoplastic progression (17). In addition, aberrant age-related as well as cancer-specific methylation of promoter-associated CpG islands of MGMT, and SFRP2 genes can lead to adenoma related neoplasia (6).
2. Objectives
Analysis methylation status of MGMT and SFRP2 genes might be particularly useful for early detection and risk assessment in patients with increased risk to develop these tumors (18). To date, little is known about the role of epigenetics in pathogenesis of polyp and adenoma in Iran. Consequently, the current study aimed to assess the methylation status of MGMT and SFRP2 genes in tissue samples of patients with polyp for early detection of polyp in Iranian patients.
3. Materials and Methods
3.1. Patients and Tumor Specimens
Of the total 54 polyp patients undergoing colonoscopic evaluation at clinical centers affiliated to Shiraz University of Medical Sciences, Southern Iran, in two years (2011 - 2013), 48 met the inclusion criteria of the research; new onset was not previously treated and histological evaluation by an expert pathologist documented clinico-pathologically in the patients with polyp; 48 polyp samples as well as corresponding normal tissues were obtained from the patients. Also 20 age- and gender-matched healthy subjects were selected from 50 volunteers who performed colonoscopy and had normal colonic mucosa. Individuals with eligible criteria who provided written informed consent were enrolled. Ethics Committee and Institutional Review Board of Shiraz University of Medical Sciences approved the study. All fresh samples were snap-frozen and stored at -70°C until processing.
3.2. Extraction of DNA
Genomic DNA was extracted from samples, as described previously (19). The study used the standard phenol/chloroform method for DNA extraction from fresh tumor samples (19).
3.3. Methylation Specific PCR
Promoter methylation status of MGMT and SFRP2 genes were determined by chemical treatment with sodium bisulfite and subsequent MSP (19). In brief, this technique uses bisulfate modification to convert unmethylated, but not methylated, cytosine to uracil. MSP utilizes this difference to specifically amplify either methylated or unmethylated DNA. Locus specific PCR primers for MGMT-B and SFRP2 genes were specifically designed for methylation specific PCR (MS-PCR) and located at each gene promoter region. The sequences, annealing temperature of each primer used for amplification, and PCR products sizes were described in Table 1. The hot-start PCR reactions were performed in a 50 µL reaction volume containing 25 pmol of sense and antisense primers, 0.2 mM/L dNTPs, and 80 μg bisulfite-modified DNA in 1 × PCR buffer provided by Taq enzyme supplier. The reaction mixture was denatured at 95°C for five minutes, after which 1.5 U Taq polymerase was added; then amplified by 40 cycles, each consisting of 30 s denaturation at 95°C, proper annealing temperature for each gene (Table 1) and 30 s polymerization at 72°C, followed by a single 10-minute extension at 72°C. The universal methylated DNA (chemicon) was used as positive control for methylated alleles of MGMT-B, and SFRP2; and DNA from normal lymphocytes was used as the negative control. Then, 10 µL of amplified PCR products were mixed with 5 µL of loading dye and electrophoresed on 2.5% agarose gel containing gel red with TBE buffer and visualized under UV illumination.
| Gene | Gene Bank Number | Annealing Temperature, °C | Product Size, bp |
|---|---|---|---|
| SFRP2 | NM_003013.2 | ||
| Primer Sequence (5′-3′) | |||
| MF: TGCGTGTTTTTATTTTCGTAGTTCGC | 59 | M: 138; U: 145 | |
| MR: CCCTAAATACCGCCGCTCGCCCG TGT | |||
| UF: GTTTTGTGTGTTTTTTATTTTTGTAGTTTGT | |||
| UR: TCCCCTAAATACCACCACTCACCCA | |||
| MGMT-B | AL355531.16 | ||
| Primer Sequence (5′-3′) | 59 | M: 127; U: 127 | |
| MF: GGTCGTTTGTACGTTCGC | |||
| MR: TAACCCTTCGACCGATACAA | |||
| UF: GTAGGTTGTTTGTATGTTTGT | |||
| UR: TAACCCTTCAACCAATACAAACC |
Primer Sequence of SFRP2 and MGMT Genes
3.4. Statistical Analysis
Depending upon the sample size, associations between clinical, biological, and genotypic features were evaluated using either the Chi-square or the Fisher's exact tests. The level of significance was P < 0.05. Potential confounding variables such as age and gender were also studied. All statistical data were analyzed by SPSS software, version 11.5 (SPSS Inc., Chicago, IL).
4. Results
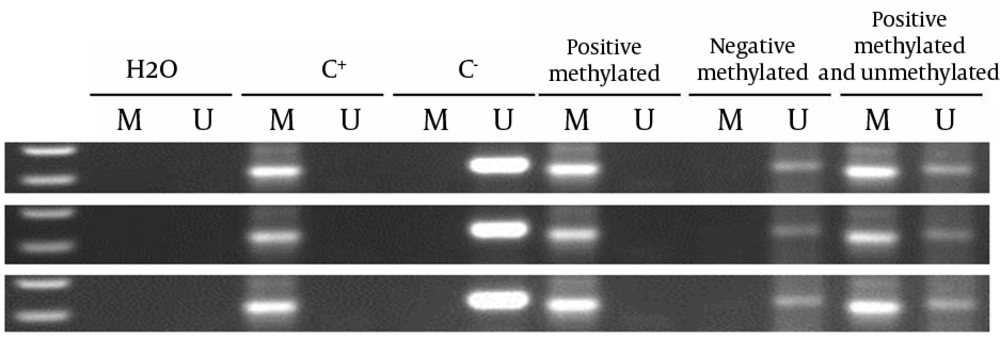
In the current study, no methylation was observed in normal controls and normal adjacent tissues of the patients with polyp for both genes. Mean and median of age of subjects and gender distribution is shown in Table 2. In the current study, 64.7% and 35.3 % of total subjects were female and male, respectively. Median and mean age of the subjects were 53 and 50.97, respectively. Total methylation level of SFRP2 and MGMT genes were 81.2% and 66.6%, respectively. According to the study, the methylation levels of SFRP2 and MGMT genes were 81.3%, 53.3% respectively; 80%, and 71% in female and male tissue samples, respectively. SFRP2 and MGMT genes methylation are shown in Figure 1. Moreover, Table 3 shows the stratification analysis of tumors and genes promoters of methylation frequencies. According to median age, the subjects were divided into two groups of ≥ 53 and < 53 years. There was no significant methylation alteration with respect to age. However, an increase trend was found in the methylation status of SFRP2 gene in individuals above 53 years old. Interestingly, the methylation level in at least one gene was 93.8% that indicated the importance of these two genes as an appropriate marker to detect polyp and adenoma. In this regard, 45.8% of the samples were methylated for both genes. Herein the methylation level of SFRP2 gene is higher than that of MGMT gene, which is indicating its key role in pathogenesis of polyp and adenoma lesions.
| Variable | Frequency |
|---|---|
| Gender, % | |
| Male | 35.3 |
| Female | 64.7 |
| Median age | 53 |
| Mean age | 60 ± 18.7 |
Distribution of the Selected Characteristics in the Subjects
The presence of a visible PCR product in those lanes marked U indicates the presence of unmethylated genes; the presence of a product in those lanes marked M indicates the presence of methylated genes. Lane 1 indicates the 50 base pair DNA size marker. Universal methylated DNA (UMD), unmethylated lymphocytes (lymphocytes) DNA and H2O were used as positive and negative controls, and NTC respectively.
| Variable | Methylation Positive Genes | ||
|---|---|---|---|
| SFRP2 | MGMT | At Least One Methylated Gene | |
| Gender | |||
| Male | 81.3 | 53.3 | 93.8 |
| Female | 80.0 | 71.0 | 93.8 |
| Total | 81.2 | 66.6 | 93.8 |
| P-value | 1.000 | 0.325 | 1.000 |
| Age | |||
| < 53 | 70.8 | 66.7 | 92.0 |
| ≥ 53 | 90.9 | 63.6 | 95.7 |
| P-value | 0.139 | 1.000 | 1.000 |
Stratification Analysis of Tumors and Genes Promoters Methylation Frequencies a
5. Discussion
The current study results showed that methylation of SFRP2 and MGMT genes plays a significant role in polyp formation. However, it seems that SFRP2, as the antagonist of Wnt signaling pathway, is more important. It is widely believed that CRC develops as a result of progressive accumulation of genetic alterations that lead to the transformation of normal epithelium to adenocarcinoma (20). In spite of genetic alterations, recent studies show that aberrant DNA methylation plays a causal role in the molecular pathogenesis of several cancers, including colon cancer (20, 21). Furthermore, the aberrant methylation of MGMT and SFRP2 genes indicates early events in colon cancer progression (22). Recent studies revealed that promoter hypermethylation in these genes is involved in progression of sporadic colon cancer (20). In addition, CpG island methylator phenotype (CIMP) is observed in proximal hyperplastic (serrated) polyps, suggesting that this lesion may be a precursor to progress colon cancer (23). The current study evaluated the methylation status of MGMT and SFRP2 genes in tissue samples of patients with polyp and adenomas (24). The findings provided support for a unique role of DNA methylation of the two genes in the formation of polyps and adenomas. The gene SFRP2 had higher methylation frequency than MGMT. In addition, the total methylation status in both genes was higher in males than females. These findings show that probably the methylation level and the prevalence of polyp development is higher in males than females, and SFRP2 is a key gene in this process. There was no significant relationship between age, and methylation level of the two genes. However, compared with MGMT gene, hypermethylation of SFRP2 in the individuals lower than 53 years old was less than those of the ones higher than 53. The study results correlate with those of the previous researches regarding the lack of significant correlation between MGMT or SFRP2 methylation and age (25). The current study results confirm several other studies on the effectiveness of monitoring patients with CRC prone disorder using these DNA markers; they also correlate with those of the previous studies regarding the higher methylated amount of SFRP2 rather than MGMT found in tissue samples (26-30). However the precise amount of methylation found in the current study was higher than those of most of the previous ones, which could be because of the racial differences of the current study subjects and those of the other studies, and limited statistical population in the current study. This amount of epigenetic changes heightens the risks of other genetic changes such as P53 and K-ras mutations, which makes it a serious field for further research and follow up in larger populations with several types of polyp. The observation should be completed in early and advanced polyps and adenomas, whereas the methylation of these genes was an early event in the formation of the polyp in the studied population. These findings imply that aberrantly methylated genes have the potential to be used as early detection markers for polyp formation and adenomas. Epigenetic alteration of Wnt antagonist and DNA repair pathways are correlated with neoplasm changes.