1. Background
Ranking as the second leading cause of death worldwide, breast cancer is a major health concern (1). Over the past decades, survival rate of breast cancer patients has improved due to advancement in breast cancer early diagnosis methods and novel treatment strategies (2). However, patient’s response rate to treatment is not satisfactory as the result of developing drug resistance (3). Multidrug Resistance (MDR) is a major challenge impairing breast cancer successful chemotherapy (1, 4). Drug resistance or ineffective chemotherapy agent administration are two major reasons for unsuccessful treatment in 90% of patients with metastatic cancer (5). Different mechanisms are attributed to the multidrug resistance phenomenon such as reduction in drug-induced apoptosis, induction of drug detoxification mechanisms and active drug efflux from cancer cells by ATP-binding cassette (ABC) transporters (4, 6, 7).
The ABC transporters are membrane proteins belonging to the ABC superfamily. The ABC superfamily is classified into seven distinct subfamilies ranging from ABCA to ABCG, on the basis of sequence homology. The ABC proteins use energy produced from ATP hydrolysis to actively transport different compounds across the cell membrane and are also involved in many diseases and malignancies (4, 8, 9). Multidrug resistance-associated protein 1 (MRP1/ABCC1) was the first member of the ABCC subfamily, which is linked to MDR in many solid tumors. This 190-kDa protein is ubiquitously present in all human tissues and transports a wide spectrum of substrates ranging from xenobiotics such as doxorubicin, taxanes, methotrexate and imatinib to endobiotics including glutathione, leukotrienes and prostaglandins (10-12). Overexpression of this protein was reported in many solid and invasive tumors including breast cancer, ovarian cancer, lung cancer and neuroblastoma. It is responsible for tumor cells resistance to anthracyclines and methotrexate chemotherapy drugs (11). In addition, several independent studies have shown that miRNAs including miR-7, miR-345 (5), miR-1291 (11, 13), miR-133a and miR-326 (14) regulate ABCC1 expression and function by targeting 3’UTR of ABCC1 mRNA, directing drug distribution in cells and cell’s sensitivity to chemotherapeutic agents.
On the other hand, all patients don’t respond similarly to the same drug. In fact, inter-individual hereditary variations in genes encoding proteins, which are involved in drug transport, metabolism and excretion, may account for individual differences in drug response (15). In the human genome project many variations were identified among populations. Single nucleotide polymorphism (SNP) is one of the most abundant type of variations present in > 1% of the population. Depending on the site of SNP (i.e. in the non-coding, coding and regulatory regions of the genes) it may have different outcomes ranging from no change in the quantity and quality of encoded proteins to change in structure and amount of cell protein product (16). There are many single nucleotide polymorphisms (SNPs) identified in ABCC1 gene sequences that are involved in drug resistance and cytotoxicity, disease susceptibility, prognosis and severity (12). The non-synonymous polymorphism G2168A (rs4148356) in exon 17 is strongly associated with reduced MRP1 transport activity leading to increased response to platinum/taxane in patients with advanced ovarian cancer (17). In 2013, Vulsteke et al. reported that variants of G2012T (rs45511401) and T825C (rs246221) non-synonymous polymorphisms correlated with hematological cytotoxicity after receiving neo-adjuvant therapy in breast cancer patients (18). In another study researchers addressed the effects of ABCC1 5’UTR G1666A polymorphism (rs4148330) on hepatocellular cancer outcome in patients. They also found that mutant genotype carriers had better outcome and more disease free survival (19). Besides, 3’UTR T866A (rs212090) polymorphism was another example of non-coding SNP, which was strongly associated with chronic obstructive pulmonary disease (COPD) severity (20) and lung cancer susceptibility (21) in two different studies. In general, genetic variations, inhibition or change in ABCC1 expression may alter drug disposition, cytotoxicity and clinical outcome (11).
C543T and G810C polymorphisms are two prevalent SNPs located at ABCC1 3’UTR with minor allele frequencies (MAF) of T = 0.29 and G = 0.48 per 1000 genomes, respectively. As, only a few independent studies have been undertaken to check the possible associations of these SNPs with lung cancer susceptibility (21), COPD severity (20) and drug’s pharmacokinetics (22, 23), their potential clinical significances are rarely understood.
2. Objectives
Considering the overexpression of MRP1 in breast carcinomas, especially after receiving chemotherapeutic agents (24-26), and contribution of several miRNAs in multidrug resistance phenomenon by targeting ABCC1 mRNA 3’UTR (5, 11, 13, 14), we analyzed if these two ABCC1 3’UTR polymorphisms are connected to breast cancer development.
3. Materials and Methods
3.1. Subjects
This study was a comparative descriptive analysis on 44 patients of Emam Khomeini hospital (Tehran, Iran), diagnosed with grade two breast cancer, receiving chemotherapeutic agents, irrespective of their molecular subtypes and clinicopathological status. Furthermore, 25 non-cancerous individuals, who visited Fardis central laboratory (Alborz, Iran), were used as the control group. Participants of the control group were females older than 35 years with no history of breast cancer. Informed consent was taken from the participants of both groups and this study was approved by ethics committee of Iran University of Medical Sciences (IUMS). The number of cases and controls were determined based on the incidence of breast cancer in Iran and allelic frequencies of each SNP, based on similar studies on Asian populations available on the dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) database.
3.2. DNA Extraction and Genotyping
This study was conducted at the biotechnology research laboratory and molecular and cellular research center of Iran University of Medical Sciences. Genomic DNA was extracted from whole blood samples using the Tiangene genomic DNA extraction kit (Cat number: DP304-02) and stored at -20°C. Nested-PCR was conducted with the outer F and outer R primers to amplify a 523 bp product. Next, one to ten (1:10) diluted 523 bp products were used as templates for Allele Specific-PCR (AS-PCR) to identify C and G alleles of G801C polymorphism. Tiangene master mix (cat number: KT205) was used for the PCR. The primers used for the Nested-PCR and AS-PCR were as follows:
Nested PCR:
Optimum Tm: 65°C; Product size: 523 bp.
Outer F: 5' GCTCCCATCACCTCTAACATCC 3'
Outer R: 5' AGCTGGTTGCTCACTCTCAGTC 3'.
AS-PCR:
Optimum Tm: 61°C; Product size: 339 bp.
Inner F (common primer): 5' TTCATTTCCTTGGGGCTGC 3'.
Inner R C (specific for C allele): 5' AAAAAAGGGAAAGCCTGGAATG 3'.
Inner R G (specific for G allele): 5' AAAAAAGGGAAAGCCTGGAATC 3'.
In the next step, we analyzed C and T alleles of C543T polymorphism using a specific restriction enzyme, StyI (Thermo Scientific cat number: Eco130I), which recognized and cut the 523 bp PCR product in the presence of allele C. During the final step, we checked different variants of C543T and G801C by sending the 523-bp PCR products of ten randomly selected samples for sequencing.
3.3. Statistical Analysis
All data are represented by frequencies, means and percentages. The possible association of participant’s alleles and genotypes with breast cancer was analyzed by the chi-square (X2) test using SPSS 15.1 software and P values of < 0.05 were considered significant. We used an on-line software (http://www.oege.org/software/hardy-weinberg.html) to evaluate if the observed genotypic frequencies correlated with the Hardy-Weinberg equilibrium (HWE) and P values of < 0.01 were considered as violation of HWE.
4. Results
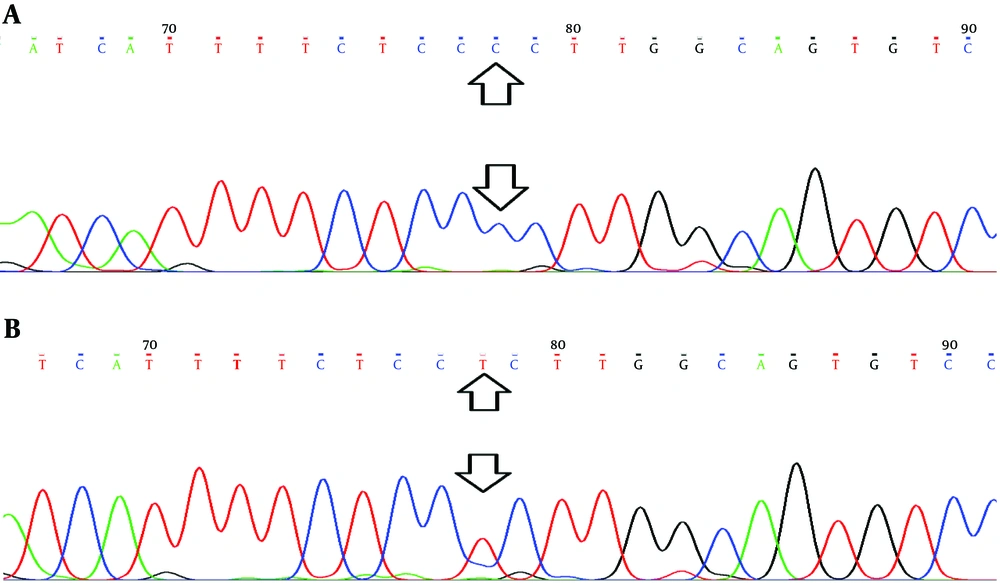
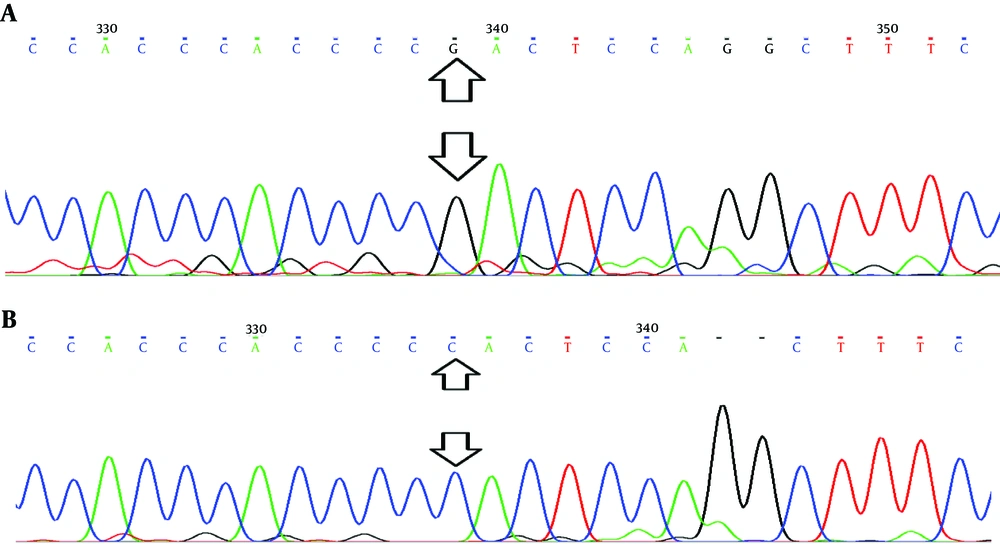
This study was conducted on 44 patients with breast cancer (cases) and 25 healthy individuals (control). The rs3743527 (C543T) allelic frequencies of C and T alleles were 56.8% and 43.2% in cases (C > T) and 64.6% and 35.4% in controls (C > T), respectively. Analysis of rs129081 (G801C) variants showed that allelic frequencies of Guanine (G) were higher than Cytosine (C) in both groups. The allelic frequencies of G allele were 56.8% in cases and 60% in controls. The C allelic frequencies were 43.2% and 40% in cases and controls, respectively. Besides, results of sequencing confirmed the presence of polymorphic variants of rs3743527 (C543T) and rs129081 (G801C) in the DNA samples (Figures 1 and 2). However, using a multiple sequence alignment algorithm, ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/), we found another polymorphic site in ten sequenced samples. Checking the NCBI SNP database (http://www.ncbi.nlm.nih.gov/), we found that this belonged to polymorphic site rs212090 (T866A) in ABCC1 3’UTR. The allelic frequencies of Adenine (A) and Thymine (T) was seven (0.7) and three (0.3) out of ten, respectively (Figure 3).
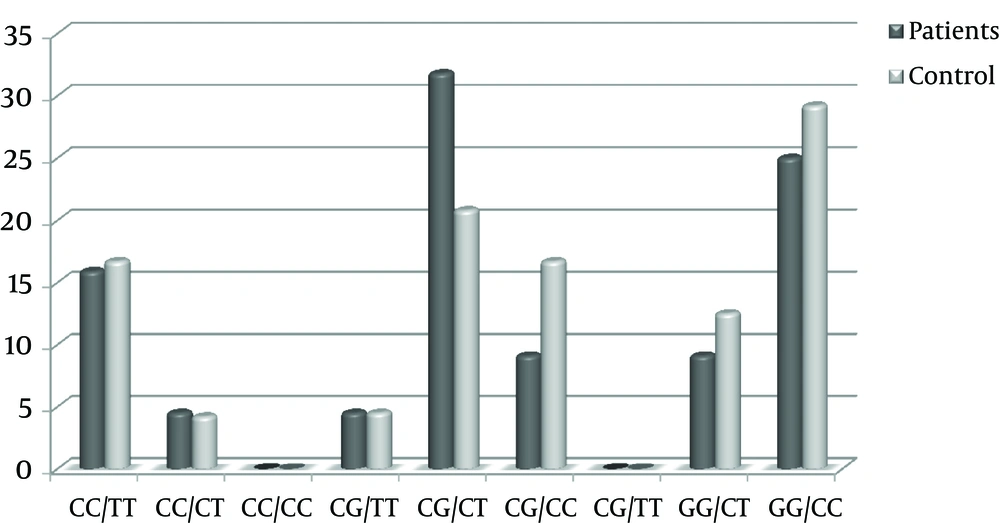
The observed C543T and G801C genotypic frequencies are shown in Table 1 didn’t deviate from expected frequencies, according to HWE in both control and case groups (P value > 0.01). Distribution of C543T and G801C genotypic frequencies didn’t significantly differ between patient and control groups, according to the X2 test analysis (P value > 0.05). The mean genotypic frequencies of G801C observed in the two groups were 20%, 37% and 43% for CC, GG and CG genotypes, respectively. Regarding the C543T genotypic frequencies, genotype CT with average frequency of 42% and genotype TT with average frequency of 19% were the highest and lowest genotypes seen in cases and controls. Yet, the results of X2 test and sequencing were indicative of a strong correlation between G801C and C543T genotypic distribution in both groups (P value = 0), i.e. on average, 27% of participants carried genotypes GG/CC, 26% genotypes CG/CT and 16% genotypes CC/TT (Figure 4).
| Nucleotide Polymorphism | Genotypes | Case | Control | P Value |
|---|---|---|---|---|
| rs129081 (G801C) | 0.87 | |||
| CC | 9 (20.5) | 5 (20%) | ||
| CG | 20 (45.5) | 10 (40%) | ||
| GG | 15 (34.1) | 10 (40%) | ||
| Total | 44 (100) | 25 (100%) | ||
| rs3743527 (C543T) | 0.63 | |||
| TT | 9 (20.5) | 4 (16.7%) | ||
| CT | 20 (45.5) | 9 (37.5%) | ||
| CC | 15 (34.1) | 11 (45.8%) | ||
| Total | 44 (100) | 24 (100%) |
aValues are expressed as No. (%).
5. Discussion
In this study the potential associations of ABCC1 3’UTR SNPs (rs3743527 and rs129081) with breast cancer susceptibility were investigated.
According to the results of this study, there were no significant differences in allelic and genotypic distribution of rs3743527 (C543T) and rs129081 (G801C) polymorphisms between breast cancer patients and healthy individuals. Similarly, the study undertaken by Coelho et al. in 2011 found no relationship between clinical and pharmacokinetics of telatinib with variants of rs129081 in patients with a solid tumor (23). Likewise, investigating the potential correlations of rs3743527 variants with lung cancer susceptibility in a Chinese population in 2009 (21) and virological failure in HIV+ Brazilian patients under anti-retroviral therapy in 2013 (22), revealed no significant associations. However, the variants of rs212090 (T866A), detected by sequencing in our study (A > T), significantly correlated with lung cancer susceptibility in a Chinese population (21).
It is important to note that in the present study analysis of these polymorphisms in a larger sample population was not possible due to time and budget limitations. Information of patient’s clinicopathological features was also unavailable, so that study of the potential relationships between patient’s molecular subtypes, response to chemotherapeutic agents and clinical outcomes was not feasible.
Altogether, allelic and genotypic distributions of rs3743527 and rs129081 variants are similar in cases and controls, so neither of them may be associated with the risk of breast cancer. Yet, regarding the potential miRNA/ABCC1 mRNA interactions, the ABCC1 contributions in multidrug resistance phenomenon as well as limitations of the present study, more comprehensive studies are required to investigate the potential relationships of rs3743527 and rs129081 variants in breast cancer patients.
5.1. Conclusion
This study may exclude rs3743527 and rs129081 variants of ABCC1 SNPs from the profile of genetic variations involved in breast cancer development; the profile which could aid scientists in early breast cancer detection and personalized cancer therapy in the near future.